A Public Database of Memory and Naive B-Cell Receptor Sequences
- PMID: 27513338
- PMCID: PMC4981401
- DOI: 10.1371/journal.pone.0160853
A Public Database of Memory and Naive B-Cell Receptor Sequences
Abstract
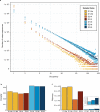
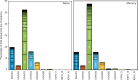
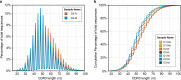
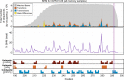
The vast diversity of B-cell receptors (BCR) and secreted antibodies enables the recognition of, and response to, a wide range of epitopes, but this diversity has also limited our understanding of humoral immunity. We present a public database of more than 37 million unique BCR sequences from three healthy adult donors that is many fold deeper than any existing resource, together with a set of online tools designed to facilitate the visualization and analysis of the annotated data. We estimate the clonal diversity of the naive and memory B-cell repertoires of healthy individuals, and provide a set of examples that illustrate the utility of the database, including several views of the basic properties of immunoglobulin heavy chain sequences, such as rearrangement length, subunit usage, and somatic hypermutation positions and dynamics.
Conflict of interest statement
Figures


References
-
- Davis MM, Calame K, Early PW, Livant DL, Joho R, Weissman IL, et al. An immunoglobulin heavy-chain gene is formed by at least two recombinational events. Nature. 1980;283(5749):733–9. Epub 1980/02/21. . - PubMed
-
- Watson CT, Steinberg KM, Huddleston J, Warren RL, Malig M, Schein J, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013;92(4):530–46. Epub 2013/04/02. 10.1016/j.ajhg.2013.03.004 - DOI - PMC - PubMed
-
- Murphy K, Travers P, Walport M. Janeway's Immunobiology. 7th ed. New York, NY: Garland Science; 2008.
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–63. Epub 2000/09/28. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

