Soluble bioactive microbial mediators regulate proteasomal degradation and autophagy to protect against inflammation-induced stress
- PMID: 27514476
- PMCID: PMC5142193
- DOI: 10.1152/ajpgi.00092.2016
Soluble bioactive microbial mediators regulate proteasomal degradation and autophagy to protect against inflammation-induced stress
Abstract
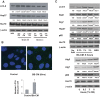
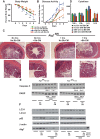
Bifidobacterium breve and other Gram-positive gut commensal microbes protect the gastrointestinal epithelium against inflammation-induced stress. However, the mechanisms whereby these bacteria accomplish this protection are poorly understood. In this study, we examined soluble factors derived from Bifidobacterium breve and their impact on the two major protein degradation systems within intestinal epithelial cells, proteasomes and autophagy. Conditioned media from gastrointestinal Gram-positive, but not Gram-negative, bacteria activated autophagy and increased expression of the autophagy proteins Atg5 and Atg7 along with the stress response protein heat shock protein 27. Specific examination of media conditioned by the Gram-positive bacterium Bifidobacterium breve (Bb-CM) showed that this microbe produces small molecules (<3 kDa) that increase expression of the autophagy proteins Atg5 and Atg7, activate autophagy, and inhibit proteasomal enzyme activity. Upregulation of autophagy by Bb-CM was mediated through MAP kinase signaling. In vitro studies using C2BBe1 cells silenced for Atg7 and in vivo studies using mice conditionally deficient in intestinal epithelial cell Atg7 showed that Bb-CM-induced cytoprotection is dependent on autophagy. Therefore, this work demonstrates that Gram-positive bacteria modify protein degradation programs within intestinal epithelial cells to promote their survival during stress. It also reveals the therapeutic potential of soluble molecules produced by these microbes for prevention and treatment of gastrointestinal disease.
Keywords: Atg5; Atg7; cell death; cell survival; host-microbe interaction; inflammatory bowel diseases; microbiota; microflora; mucosal inflammation; peptidoglycan.
Copyright © 2016 the American Physiological Society.
Figures




References
-
- Adolph TE, Tomczak MF, Niederreiter L, Ko HJ, Bock J, Martinez-Naves E, Glickman JN, Tscurtschenthaler M, Hartwig J, Hosomi S, Fiak MB, Cusick JL, Kohno K, Iwawaki T, Billman-Born S, Raine T, Bharti R, Lucius R, Kweon MN, Marciniak SJ, Choi A, Hagen SJ, Schreiber S, Rosensteil P, Kaser A, Blumberg RS. Paneth cells as a site of origin for intestinal inflammation. Nature 503: 272–276, 2013. - PMC - PubMed
-
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500: 232–236, 2013. - PubMed
-
- Basler M, Dajee M, Moll C, Groettrup M, Kirk CJ. Prevention of experimental colitis by a selective inhibitor of the immunoproteasome. J Immunol 185: 634–641, 2010. - PubMed
-
- Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol 34: 286–297, 2012. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

