A new survivin tracer tracks, delocalizes and captures endogenous survivin at different subcellular locations and in distinct organelles
- PMID: 27514728
- PMCID: PMC4981888
- DOI: 10.1038/srep31177
A new survivin tracer tracks, delocalizes and captures endogenous survivin at different subcellular locations and in distinct organelles
Abstract
Survivin, the smallest member of the inhibitor of apoptosis protein family, plays a central role during mitosis and exerts a cytoprotective function. Survivin is highly expressed in most cancer types and contributes to multiple facets of carcinogenesis. The molecular mechanisms underlying its highly diverse functions need to be extensively explored, which is crucial for rational design of future personalized therapeutics. In this study, we have generated an alpaca survivin nanobody (SVVNb8) that binds with low nanomolar affinity to its target. When expressed as an intrabody in HeLa cells, SVVNb8 faithfully tracks survivin during different phases of mitosis without interfering with survivin function. Furthermore, coupling SVVNb8 with a subcellular delocalization tag efficiently redirects endogenous survivin towards the nucleus, the cytoplasm, peroxisomes and even to the intermembrane space of mitochondria where it presumably interacts with resident mitochondrial survivin. Based on our findings, we believe that SVVNb8 is an excellent instrument to further elucidate survivin biology and topography, and can serve as a model system to investigate mitochondrial and peroxisomal (survivin) protein import.
Figures
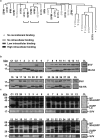

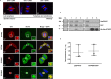
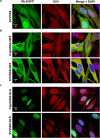

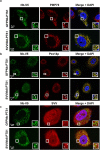

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

