Functional and Structural Characterization of Novel Type of Linker Connecting Capsid and Nucleocapsid Protein Domains in Murine Leukemia Virus
- PMID: 27514744
- PMCID: PMC5034055
- DOI: 10.1074/jbc.M116.746461
Functional and Structural Characterization of Novel Type of Linker Connecting Capsid and Nucleocapsid Protein Domains in Murine Leukemia Virus
Abstract
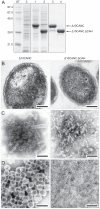
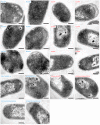
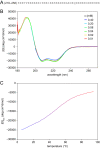
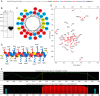
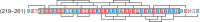
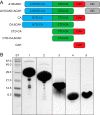
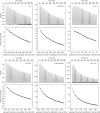
The assembly of immature retroviral particles is initiated in the cytoplasm by the binding of the structural polyprotein precursor Gag with viral genomic RNA. The protein interactions necessary for assembly are mediated predominantly by the capsid (CA) and nucleocapsid (NC) domains, which have conserved structures. In contrast, the structural arrangement of the CA-NC connecting region differs between retroviral species. In HIV-1 and Rous sarcoma virus, this region forms a rod-like structure that separates the CA and NC domains, whereas in Mason-Pfizer monkey virus, this region is densely packed, thus holding the CA and NC domains in close proximity. Interestingly, the sequence connecting the CA and NC domains in gammaretroviruses, such as murine leukemia virus (MLV), is unique. The sequence is called a charged assembly helix (CAH) due to a high number of positively and negatively charged residues. Although both computational and deletion analyses suggested that the MLV CAH forms a helical conformation, no structural or biochemical data supporting this hypothesis have been published. Using an in vitro assembly assay, alanine scanning mutagenesis, and biophysical techniques (circular dichroism, NMR, microcalorimetry, and electrophoretic mobility shift assay), we have characterized the structure and function of the MLV CAH. We provide experimental evidence that the MLV CAH belongs to a group of charged, E(R/K)-rich, single α-helices. This is the first single α-helix motif identified in viral proteins.
Keywords: capsid protein (CA); charged assembly helix (CAH); circular dichroism (CD); electron microscopy (EM); murine leukemia virus (MLV); nuclear magnetic resonance (NMR); retrovirus; single alpha-helix (SAH); spacer peptide (SP); virus assembly.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures

Similar articles
-
Structure and architecture of immature and mature murine leukemia virus capsids.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11751-E11760. doi: 10.1073/pnas.1811580115. Epub 2018 Nov 26. Proc Natl Acad Sci U S A. 2018. PMID: 30478053 Free PMC article.
-
Structural Determinants of Virion Assembly and Release in the C Terminus of the Mason-Pfizer Monkey Virus Capsid Protein.J Virol. 2021 Sep 9;95(19):e0061521. doi: 10.1128/JVI.00615-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287037 Free PMC article.
-
Role of Mason-Pfizer monkey virus CA-NC spacer peptide-like domain in assembly of immature particles.J Virol. 2014 Dec;88(24):14148-60. doi: 10.1128/JVI.02286-14. Epub 2014 Oct 1. J Virol. 2014. PMID: 25275119 Free PMC article.
-
Nucleic Acid Binding by Mason-Pfizer Monkey Virus CA Promotes Virus Assembly and Genome Packaging.J Virol. 2016 Apr 14;90(9):4593-4603. doi: 10.1128/JVI.03197-15. Print 2016 May. J Virol. 2016. PMID: 26912613 Free PMC article.
-
Mutations in the spacer peptide and adjoining sequences in Rous sarcoma virus Gag lead to tubular budding.J Virol. 2008 Jul;82(14):6788-97. doi: 10.1128/JVI.00213-08. Epub 2008 Apr 30. J Virol. 2008. PMID: 18448521 Free PMC article.
Cited by
-
Structure and architecture of immature and mature murine leukemia virus capsids.Proc Natl Acad Sci U S A. 2018 Dec 11;115(50):E11751-E11760. doi: 10.1073/pnas.1811580115. Epub 2018 Nov 26. Proc Natl Acad Sci U S A. 2018. PMID: 30478053 Free PMC article.
-
ER/K-link-Leveraging a native protein linker to probe dynamic cellular interactions.Methods Enzymol. 2021;647:173-208. doi: 10.1016/bs.mie.2020.10.002. Epub 2020 Nov 18. Methods Enzymol. 2021. PMID: 33482988 Free PMC article. Review.
-
Effect of Small Polyanions on In Vitro Assembly of Selected Members of Alpha-, Beta- and Gammaretroviruses.Viruses. 2021 Jan 18;13(1):129. doi: 10.3390/v13010129. Viruses. 2021. PMID: 33477490 Free PMC article.
-
In vitro methods for testing antiviral drugs.Biotechnol Adv. 2018 May-Jun;36(3):557-576. doi: 10.1016/j.biotechadv.2017.12.016. Epub 2017 Dec 29. Biotechnol Adv. 2018. PMID: 29292156 Free PMC article. Review.
-
Structural Determinants of Virion Assembly and Release in the C Terminus of the Mason-Pfizer Monkey Virus Capsid Protein.J Virol. 2021 Sep 9;95(19):e0061521. doi: 10.1128/JVI.00615-21. Epub 2021 Jul 21. J Virol. 2021. PMID: 34287037 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

