COMAN: a web server for comprehensive metatranscriptomics analysis
- PMID: 27515514
- PMCID: PMC4982211
- DOI: 10.1186/s12864-016-2964-z
COMAN: a web server for comprehensive metatranscriptomics analysis
Abstract
Background: Microbiota-oriented studies based on metagenomic or metatranscriptomic sequencing have revolutionised our understanding on microbial ecology and the roles of both clinical and environmental microbes. The analysis of massive metatranscriptomic data requires extensive computational resources, a collection of bioinformatics tools and expertise in programming.
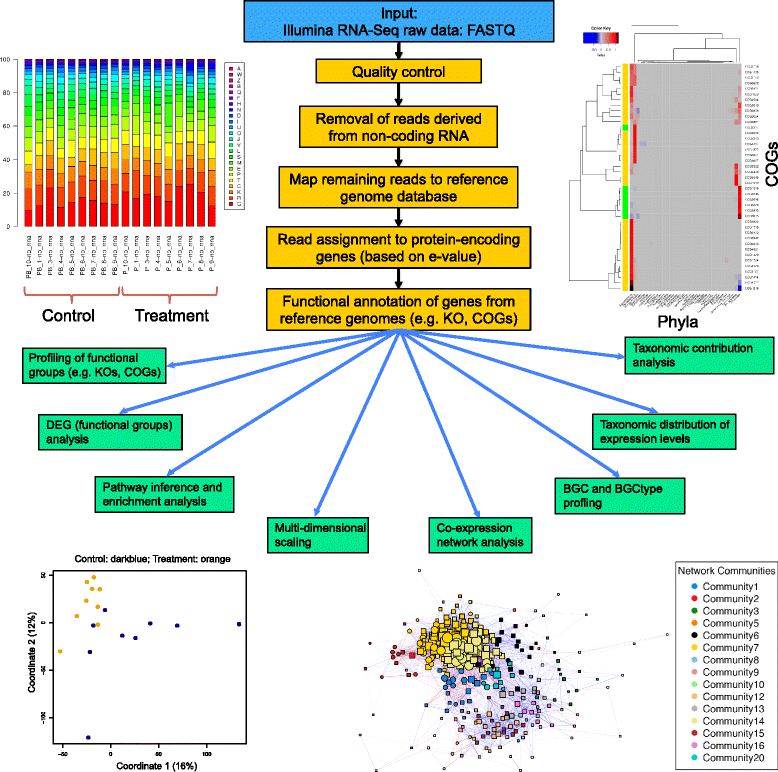
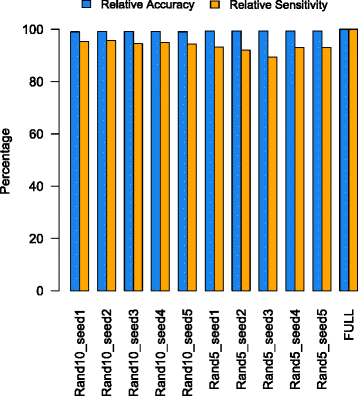
Results: We developed COMAN (Comprehensive Metatranscriptomics Analysis), a web-based tool dedicated to automatically and comprehensively analysing metatranscriptomic data. COMAN pipeline includes quality control of raw reads, removal of reads derived from non-coding RNA, followed by functional annotation, comparative statistical analysis, pathway enrichment analysis, co-expression network analysis and high-quality visualisation. The essential data generated by COMAN are also provided in tabular format for additional analysis and integration with other software. The web server has an easy-to-use interface and detailed instructions, and is freely available at http://sbb.hku.hk/COMAN/ CONCLUSIONS: COMAN is an integrated web server dedicated to comprehensive functional analysis of metatranscriptomic data, translating massive amount of reads to data tables and high-standard figures. It is expected to facilitate the researchers with less expertise in bioinformatics in answering microbiota-related biological questions and to increase the accessibility and interpretation of microbiota RNA-Seq data.
Keywords: Computational biology; Metatranscriptomics; Microbial RNA-Seq; Microbial community; Web servers.
Figures
References
-
- Booijink CC, Boekhorst J, Zoetendal EG, Smidt H, Kleerebezem M, de Vos WM. Metatranscriptome analysis of the human fecal microbiota reveals subject-specific expression profiles, with genes encoding proteins involved in carbohydrate metabolism being dominantly expressed. Appl Environ Microbiol. 2010;76(16):5533–5540. doi: 10.1128/AEM.00502-10. - DOI - PMC - PubMed
-
- Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, et al. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A. 2010;107(16):7503–7508. doi: 10.1073/pnas.1002355107. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

