Estimating species richness using environmental DNA
- PMID: 27516876
- PMCID: PMC4972244
- DOI: 10.1002/ece3.2186
Estimating species richness using environmental DNA
Abstract
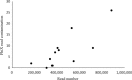
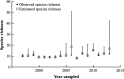
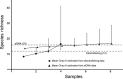
The foundation for any ecological study and for the effective management of biodiversity in natural systems requires knowing what species are present in an ecosystem. We assessed fish communities in a stream using two methods, depletion-based electrofishing and environmental DNA metabarcoding (eDNA) from water samples, to test the hypothesis that eDNA provides an alternative means of determining species richness and species identities for a natural ecosystem. In a northern Indiana stream, electrofishing yielded a direct estimate of 12 species and a mean estimated richness (Chao II estimator) of 16.6 species with a 95% confidence interval from 12.8 to 42.2. eDNA sampling detected an additional four species, congruent with the mean Chao II estimate from electrofishing. This increased detection rate for fish species between methods suggests that eDNA sampling can enhance estimation of fish fauna in flowing waters while having minimal sampling impacts on fish and their habitat. Modern genetic approaches therefore have the potential to transform our ability to build a more complete list of species for ecological investigations and inform management of aquatic ecosystems.
Keywords: Chao estimator; electrofishing; freshwater community; metabarcoding; species identity.
Figures


References
-
- Adamson, E. A. , and Hurwood D. A.. 2016. Molecular ecology and stock identification. pp. 811–829 in Craig J. F., ed. freshwater fish ecologyl. John Wiley & Sons, Ltd, Chichester, UK.
-
- Arponen, A. , Heikkinen R. K., Thomas C. D., and Moilanen A.. 2005. The value of biodiversity in reserve selection: representation, species weighting, and benefit functions. Conserv. Biol. 19:2009–2014.
-
- Barnes, M. A. , Turner C. R., Jerde C. L., Renshaw M. A., Chadderton W. L., and Lodge D. M.. 2014. Environmental conditions influence eDNA persistence in aquatic systems. Environ. Sci. Technol. 48:1819–1827. - PubMed
-
- Boyer, F. , Mercier C., Bonin A., Le Bras Y., Taberlet P., and Coissac E.. 2015. obitools: a unix‐inspired software package for DNA metabarcoding. Mol. Ecol. Resour. 16:176–182. doi:10.1111/1755‐0998.12428. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

