Active Center Control of Termination by RNA Polymerase III and tRNA Gene Transcription Levels In Vivo
- PMID: 27518095
- PMCID: PMC4982682
- DOI: 10.1371/journal.pgen.1006253
Active Center Control of Termination by RNA Polymerase III and tRNA Gene Transcription Levels In Vivo
Abstract
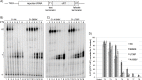
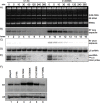
The ability of RNA polymerase (RNAP) III to efficiently recycle from termination to reinitiation is critical for abundant tRNA production during cellular proliferation, development and cancer. Yet understanding of the unique termination mechanisms used by RNAP III is incomplete, as is its link to high transcription output. We used two tRNA-mediated suppression systems to screen for Rpc1 mutants with gain- and loss- of termination phenotypes in S. pombe. 122 point mutation mutants were mapped to a recently solved 3.9 Å structure of yeast RNAP III elongation complex (EC); they cluster in the active center bridge helix and trigger loop, as well as the pore and funnel, the latter of which indicate involvement of the RNA cleavage domain of the C11 subunit in termination. Purified RNAP III from a readthrough (RT) mutant exhibits increased elongation rate. The data strongly support a kinetic coupling model in which elongation rate is inversely related to termination efficiency. The mutants exhibit good correlations of terminator RT in vitro and in vivo, and surprisingly, amounts of transcription in vivo. Because assessing in vivo transcription can be confounded by various parameters, we used a tRNA reporter with a processing defect and a strong terminator. By ruling out differences in RNA decay rates, the data indicate that mutants with the RT phenotype synthesize more RNA than wild type cells, and than can be accounted for by their increased elongation rate. Finally, increased activity by the mutants appears unrelated to the RNAP III repressor, Maf1. The results show that the mobile elements of the RNAP III active center, including C11, are key determinants of termination, and that some of the mutations activate RNAP III for overall transcription. Similar mutations in spontaneous cancer suggest this as an unforeseen mechanism of RNAP III activation in disease.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








Similar articles
-
RNA polymerase III mutants in TFIIFα-like C37 that cause terminator readthrough with no decrease in transcription output.Nucleic Acids Res. 2013 Jan 7;41(1):139-55. doi: 10.1093/nar/gks985. Epub 2012 Oct 23. Nucleic Acids Res. 2013. PMID: 23093604 Free PMC article.
-
Point mutations in the Rpb9-homologous domain of Rpc11 that impair transcription termination by RNA polymerase III.Nucleic Acids Res. 2011 Aug;39(14):6100-13. doi: 10.1093/nar/gkr182. Epub 2011 Mar 30. Nucleic Acids Res. 2011. PMID: 21450810 Free PMC article.
-
RNA Polymerase III Output Is Functionally Linked to tRNA Dimethyl-G26 Modification.PLoS Genet. 2015 Dec 31;11(12):e1005671. doi: 10.1371/journal.pgen.1005671. eCollection 2015 Dec. PLoS Genet. 2015. PMID: 26720005 Free PMC article.
-
Interactions between RNAP III transcription machinery and tRNA processing factors.Biochim Biophys Acta Gene Regul Mech. 2018 Apr;1861(4):354-360. doi: 10.1016/j.bbagrm.2018.02.003. Epub 2018 Feb 9. Biochim Biophys Acta Gene Regul Mech. 2018. PMID: 29428193 Review.
-
RNA Polymerase III Advances: Structural and tRNA Functional Views.Trends Biochem Sci. 2016 Jun;41(6):546-559. doi: 10.1016/j.tibs.2016.03.003. Epub 2016 Apr 8. Trends Biochem Sci. 2016. PMID: 27068803 Free PMC article. Review.
Cited by
-
The nuclear and cytoplasmic activities of RNA polymerase III, and an evolving transcriptome for surveillance.Nucleic Acids Res. 2021 Dec 2;49(21):12017-12034. doi: 10.1093/nar/gkab1145. Nucleic Acids Res. 2021. PMID: 34850129 Free PMC article.
-
Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases.Nat Rev Mol Cell Biol. 2022 Sep;23(9):603-622. doi: 10.1038/s41580-022-00476-9. Epub 2022 May 3. Nat Rev Mol Cell Biol. 2022. PMID: 35505252 Review.
-
A Novel Assay for RNA Polymerase I Transcription Elongation Sheds Light on the Evolutionary Divergence of Eukaryotic RNA Polymerases.Biochemistry. 2019 Apr 23;58(16):2116-2124. doi: 10.1021/acs.biochem.8b01256. Epub 2019 Apr 5. Biochemistry. 2019. PMID: 30912638 Free PMC article.
-
A structural perspective of human RNA polymerase III.RNA Biol. 2022;19(1):246-255. doi: 10.1080/15476286.2021.2022293. Epub 2021 Dec 31. RNA Biol. 2022. PMID: 35133940 Free PMC article.
-
Transcription-dependent enrichment of the yeast FACT complex influences nucleosome dynamics on the RNA polymerase III-transcribed genes.RNA. 2021 Feb 16;27(3):273-290. doi: 10.1261/rna.077974.120. RNA. 2021. PMID: 33277439 Free PMC article.
References
-
- Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292(5523):1876–82. . - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

