Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death
- PMID: 27518435
- PMCID: PMC5260504
- DOI: 10.1038/cdd.2016.78
Coordinated ubiquitination and phosphorylation of RIP1 regulates necroptotic cell death
Abstract
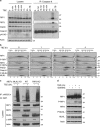
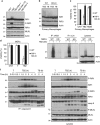
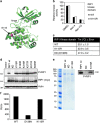
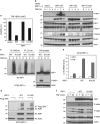
Proper regulation of cell death signaling is crucial for the maintenance of homeostasis and prevention of disease. A caspase-independent regulated form of cell death called necroptosis is rapidly emerging as an important mediator of a number of human pathologies including inflammatory bowel disease and ischemia-reperfusion organ injury. Activation of necroptotic signaling through TNF signaling or organ injury leads to the activation of kinases receptor-interacting protein kinases 1 and 3 (RIP1 and RIP3) and culminates in inflammatory cell death. We found that, in addition to phosphorylation, necroptotic cell death is regulated by ubiquitination of RIP1 in the necrosome. Necroptotic RIP1 ubiquitination requires RIP1 kinase activity, but not necroptotic mediators RIP3 and MLKL (mixed lineage kinase-like). Using immunoaffinity enrichment and mass spectrometry, we profiled numerous ubiquitination events on RIP1 that are triggered during necroptotic signaling. Mutation of a necroptosis-related ubiquitination site on RIP1 reduced necroptotic cell death and RIP1 ubiquitination and phosphorylation, and disrupted the assembly of RIP1 and RIP3 in the necrosome, suggesting that necroptotic RIP1 ubiquitination is important for maintaining RIP1 kinase activity in the necrosome complex. We also observed RIP1 ubiquitination in injured kidneys consistent with a physiological role of RIP1 ubiquitination in ischemia-reperfusion disease. Taken together, these data reveal that coordinated and interdependent RIP1 phosphorylation and ubiquitination within the necroptotic complex regulate necroptotic signaling and cell death.
Conflict of interest statement
MCdA and MK are former and all other authors are current employees of Genentech. The authors declare no conflict of interest.
Figures


References
-
- Steller H. Mechanisms and genes of cellular suicide. Science 1995; 267: 1445–1449. - PubMed
-
- Linkermann A, Stockwell BR, Krautwald S, Anders HJ. Regulated cell death and inflammation: an auto-amplification loop causes organ failure. Nat Rev Immunol 2014; 14: 759–767. - PubMed
-
- Salvesen GS, Abrams JM. Caspase activation – stepping on the gas or releasing the brakes? Lessons from humans and flies. Oncogene 2004; 23: 2774–2784. - PubMed
-
- Newton K. RIPK1 and RIPK3: critical regulators of inflammation and cell death. Trends Cell Biol 2015; 25: 347–353. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

