Dual Chromatin and Cytoskeletal Remodeling by SETD2
- PMID: 27518565
- PMCID: PMC5101839
- DOI: 10.1016/j.cell.2016.07.005
Dual Chromatin and Cytoskeletal Remodeling by SETD2
Abstract
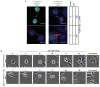
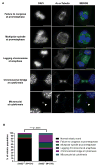
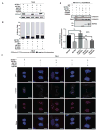
Posttranslational modifications (PTMs) of tubulin specify microtubules for specialized cellular functions and comprise what is termed a "tubulin code." PTMs of histones comprise an analogous "histone code," although the "readers, writers, and erasers" of the cytoskeleton and epigenome have heretofore been distinct. We show that methylation is a PTM of dynamic microtubules and that the histone methyltransferase SET-domain-containing 2 (SETD2), which is responsible for H3 lysine 36 trimethylation (H3K36me3) of histones, also methylates α-tubulin at lysine 40, the same lysine that is marked by acetylation on microtubules. Methylation of microtubules occurs during mitosis and cytokinesis and can be ablated by SETD2 deletion, which causes mitotic spindle and cytokinesis defects, micronuclei, and polyploidy. These data now identify SETD2 as a dual-function methyltransferase for both chromatin and the cytoskeleton and show a requirement for methylation in maintenance of genomic stability and the integrity of both the tubulin and histone codes.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures




Comment in
-
Post-translational modifications: Extension of the tubulin code.Nat Rev Mol Cell Biol. 2016 Oct;17(10):609. doi: 10.1038/nrm.2016.117. Epub 2016 Aug 24. Nat Rev Mol Cell Biol. 2016. PMID: 27552972 No abstract available.
References
-
- Agromayor M, Martin-Serrano J. Knowing when to cut and run: mechanisms that control cytokinetic abscission. Trends in cell biology. 2013;23:433–441. - PubMed
-
- Bi M, Zhao S, Said JW, Merino MJ, Adeniran AJ, Xie Z, Nawaf CB, Choi J, Belldegrun AS, Pantuck AJ, et al. Genomic characterization of sarcomatoid transformation in clear cell renal cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:2170–2175. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA198482/CA/NCI NIH HHS/United States
- R01 ES023206/ES/NIEHS NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA203012/CA/NCI NIH HHS/United States
- RC2 ES018789/ES/NIEHS NIH HHS/United States
- T32 CA009156/CA/NCI NIH HHS/United States
- K12 CA090628/CA/NCI NIH HHS/United States
- R01 CA166447/CA/NCI NIH HHS/United States
- R01 GM070862/GM/NIGMS NIH HHS/United States
- R01 ES008263/ES/NIEHS NIH HHS/United States
- F30 CA192643/CA/NCI NIH HHS/United States
- P30 ES023512/ES/NIEHS NIH HHS/United States
- T32 GM008719/GM/NIGMS NIH HHS/United States
- R01 CA121052/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

