Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes
- PMID: 27519264
- PMCID: PMC4983049
- DOI: 10.1186/s12864-016-2865-1
Regulatory complexity revealed by integrated cytological and RNA-seq analyses of meiotic substages in mouse spermatocytes
Abstract
Background: The continuous and non-synchronous nature of postnatal male germ-cell development has impeded stage-specific resolution of molecular events of mammalian meiotic prophase in the testis. Here the juvenile onset of spermatogenesis in mice is analyzed by combining cytological and transcriptomic data in a novel computational analysis that allows decomposition of the transcriptional programs of spermatogonia and meiotic prophase substages.
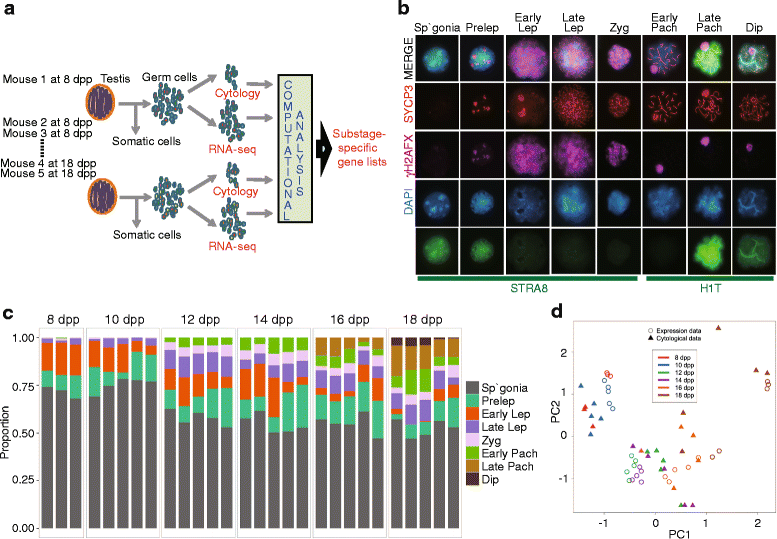
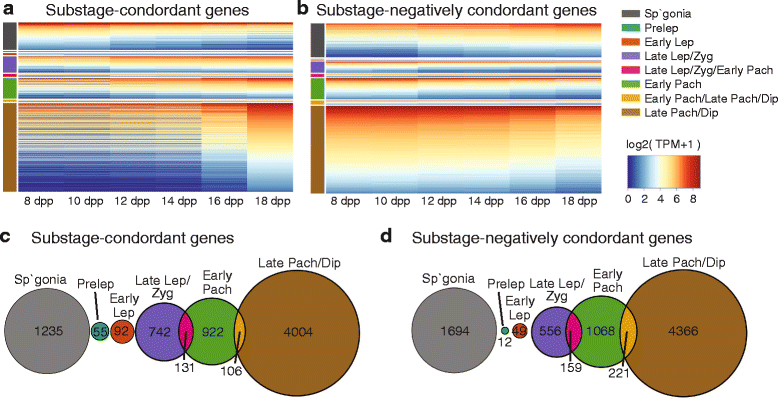
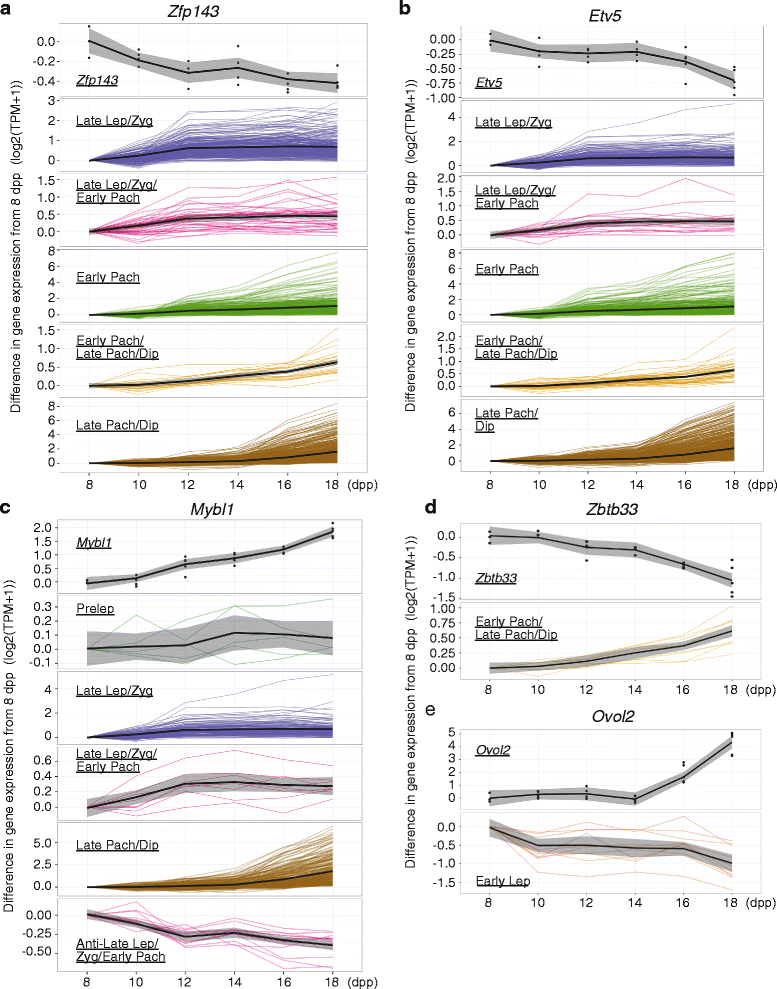
Results: Germ cells from testes of individual mice were obtained at two-day intervals from 8 to 18 days post-partum (dpp), prepared as surface-spread chromatin and immunolabeled for meiotic stage-specific protein markers (STRA8, SYCP3, phosphorylated H2AFX, and HISTH1T). Eight stages were discriminated cytologically by combinatorial antibody labeling, and RNA-seq was performed on the same samples. Independent principal component analyses of cytological and transcriptomic data yielded similar patterns for both data types, providing strong evidence for substage-specific gene expression signatures. A novel permutation-based maximum covariance analysis (PMCA) was developed to map co-expressed transcripts to one or more of the eight meiotic prophase substages, thereby linking distinct molecular programs to cytologically defined cell states. Expression of meiosis-specific genes is not substage-limited, suggesting regulation of substage transitions at other levels.
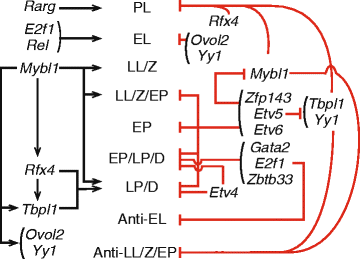
Conclusions: This integrated analysis provides a general method for resolving complex cell populations. Here it revealed not only features of meiotic substage-specific gene expression, but also a network of substage-specific transcription factors and relationships to potential target genes.
Keywords: Maximum covariance analysis; Meiosis; Mouse; RNA-seq; Spermatogenesis; Transcriptome.
Figures

References
-
- da Cruz I, Rodriguez-Casuriaga R, Santinaque FF, Farias J, Curti G, Capoano CA, Folle GA, Benavente R, Sotelo-Silveira JR, Geisinger A. Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics. 2016;17(1):294. doi: 10.1186/s12864-016-2618-1. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

