p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1
- PMID: 27519799
- PMCID: PMC5024628
- DOI: 10.1073/pnas.1603435113
p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1
Abstract
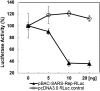
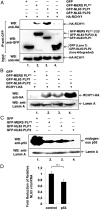
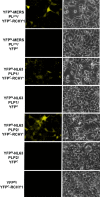
Highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) has developed strategies to inhibit host immune recognition. We identify cellular E3 ubiquitin ligase ring-finger and CHY zinc-finger domain-containing 1 (RCHY1) as an interacting partner of the viral SARS-unique domain (SUD) and papain-like protease (PL(pro)), and, as a consequence, the involvement of cellular p53 as antagonist of coronaviral replication. Residues 95-144 of RCHY1 and 389-652 of SUD (SUD-NM) subdomains are crucial for interaction. Association with SUD increases the stability of RCHY1 and augments RCHY1-mediated ubiquitination as well as degradation of p53. The calcium/calmodulin-dependent protein kinase II delta (CAMK2D), which normally influences RCHY1 stability by phosphorylation, also binds to SUD. In vivo phosphorylation shows that SUD does not regulate phosphorylation of RCHY1 via CAMK2D. Similarly to SUD, the PL(pro)s from SARS-CoV, MERS-CoV, and HCoV-NL63 physically interact with and stabilize RCHY1, and thus trigger degradation of endogenous p53. The SARS-CoV papain-like protease is encoded next to SUD within nonstructural protein 3. A SUD-PL(pro) fusion interacts with RCHY1 more intensively and causes stronger p53 degradation than SARS-CoV PL(pro) alone. We show that p53 inhibits replication of infectious SARS-CoV as well as of replicons and human coronavirus NL63. Hence, human coronaviruses antagonize the viral inhibitor p53 via stabilizing RCHY1 and promoting RCHY1-mediated p53 degradation. SUD functions as an enhancer to strengthen interaction between RCHY1 and nonstructural protein 3, leading to a further increase in in p53 degradation. The significance of these findings is that down-regulation of p53 as a major player in antiviral innate immunity provides a long-sought explanation for delayed activities of respective genes.
Keywords: E3 ubiquitin ligase RCHY1; SARS-CoV SUD; coronavirus replication; p53 antiviral activity; papain-like protease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

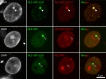



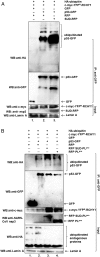


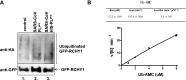
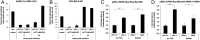

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

