Towards an automated analysis of bacterial peptidoglycan structure
- PMID: 27520322
- PMCID: PMC5203844
- DOI: 10.1007/s00216-016-9857-5
Towards an automated analysis of bacterial peptidoglycan structure
Abstract
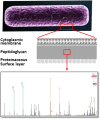
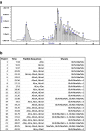
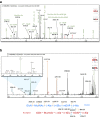
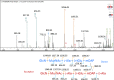
Peptidoglycan (PG) is an essential component of the bacterial cell envelope. This macromolecule consists of glycan chains alternating N-acetylglucosamine and N-acetylmuramic acid, cross-linked by short peptides containing nonstandard amino acids. Structural analysis of PG usually involves enzymatic digestion of glycan strands and separation of disaccharide peptides by reversed-phase HPLC followed by collection of individual peaks for MALDI-TOF and/or tandem mass spectrometry. Here, we report a novel strategy using shotgun proteomics techniques for a systematic and unbiased structural analysis of PG using high-resolution mass spectrometry and automated analysis of HCD and ETD fragmentation spectra with the Byonic software. Using the PG of the nosocomial pathogen Clostridium difficile as a proof of concept, we show that this high-throughput approach allows the identification of all PG monomers and dimers previously described, leaving only disambiguation of 3-3 and 4-3 cross-linking as a manual step. Our analysis confirms previous findings that C. difficile peptidoglycans include mainly deacetylated N-acetylglucosamine residues and 3-3 cross-links. The analysis also revealed a number of low abundance muropeptides with peptide sequences not previously reported. Graphical Abstract The bacterial cell envelope includes plasma membrane, peptidoglycan, and surface layer. Peptidoglycan is unique to bacteria and the target of the most important antibiotics; here it is analyzed by mass spectrometry.
Keywords: Cell wall; Cross-link; Glycoproteomics; Muropeptides; Peptidoglycan; Proteomics; Tandem mass spectrometry.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

