Use of canonical discriminant analysis to study signatures of selection in cattle
- PMID: 27521154
- PMCID: PMC4983034
- DOI: 10.1186/s12711-016-0236-7
Use of canonical discriminant analysis to study signatures of selection in cattle
Abstract
Background: Cattle include a large number of breeds that are characterized by marked phenotypic differences and thus constitute a valuable model to study genome evolution in response to processes such as selection and domestication. Detection of "signatures of selection" is a useful approach to study the evolutionary pressures experienced throughout history. In the present study, signatures of selection were investigated in five cattle breeds farmed in Italy using a multivariate approach.
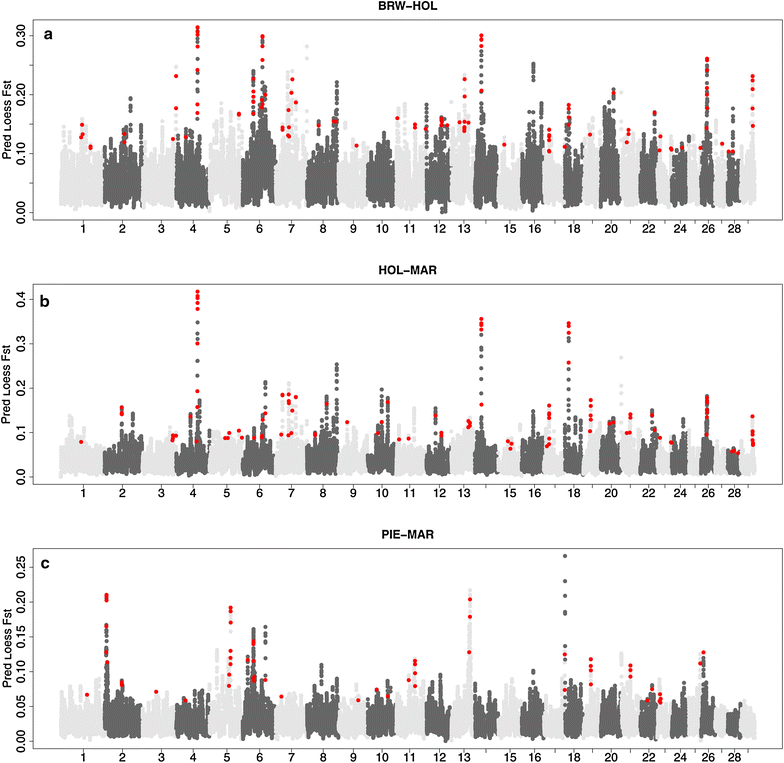
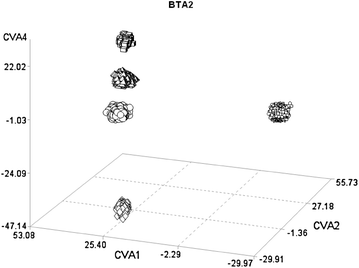
Methods: A total of 4094 bulls from five breeds with different production aptitudes (two dairy breeds: Italian Holstein and Italian Brown Swiss; two beef breeds: Piemontese and Marchigiana; and one dual purpose breed: Italian Simmental) were genotyped using the Illumina BovineSNP50 v.1 beadchip. Canonical discriminant analysis was carried out on the matrix of single nucleotide polymorphisms (SNP) genotyping data, separately for each chromosome. Scores for each canonical variable were calculated and then plotted in the canonical space to quantify the distance between breeds. SNPs for which the correlation with the canonical variable was in the 99th percentile for a specific chromosome were considered to be significantly associated with that variable. Results were compared with those obtained using an FST-based approach.
Results: Based on the results of the canonical discriminant analysis, a large number of signatures of selection were detected, among which several had strong signals in genomic regions that harbour genes known to have an impact on production and morphological bovine traits, including MSTN, LCT, GHR, SCD, NCAPG, KIT, and ASIP. Moreover, new putative candidate genes were identified, such as GCK, B3GALNT1, MGAT1, GALNTL1, PRNP, and PRND. Similar results were obtained with the FST-based approach.
Conclusions: The use of canonical discriminant analysis on 50 K SNP genotypes allowed the extraction of new variables that maximize the separation between breeds. This approach is quite straightforward, it can compare more than two groups simultaneously, and relative distances between breeds can be visualized. The genes that were highlighted in the canonical discriminant analysis were in concordance with those obtained using the FST index.
Figures



References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

