How cooperative are protein folding and unfolding transitions?
- PMID: 27522064
- PMCID: PMC5079258
- DOI: 10.1002/pro.3015
How cooperative are protein folding and unfolding transitions?
Abstract
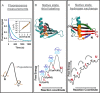
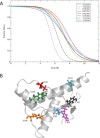
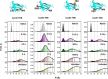
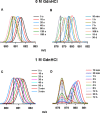
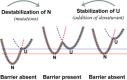
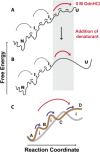
A thermodynamically and kinetically simple picture of protein folding envisages only two states, native (N) and unfolded (U), separated by a single activation free energy barrier, and interconverting by cooperative two-state transitions. The folding/unfolding transitions of many proteins occur, however, in multiple discrete steps associated with the formation of intermediates, which is indicative of reduced cooperativity. Furthermore, much advancement in experimental and computational approaches has demonstrated entirely non-cooperative (gradual) transitions via a continuum of states and a multitude of small energetic barriers between the N and U states of some proteins. These findings have been instrumental towards providing a structural rationale for cooperative versus noncooperative transitions, based on the coupling between interaction networks in proteins. The cooperativity inherent in a folding/unfolding reaction appears to be context dependent, and can be tuned via experimental conditions which change the stabilities of N and U. The evolution of cooperativity in protein folding transitions is linked closely to the evolution of function as well as the aggregation propensity of the protein. A large activation energy barrier in a fully cooperative transition can provide the kinetic control required to prevent the accumulation of partially unfolded forms, which may promote aggregation. Nevertheless, increasing evidence for barrier-less "downhill" folding, as well as for continuous "uphill" unfolding transitions, indicate that gradual non-cooperative processes may be ubiquitous features on the free energy landscape of protein folding.
Keywords: cooperativity; downhill folding; intermediates; one-state; population distributions; uphill unfolding.
© 2016 The Protein Society.
Figures

Similar articles
-
Tuning Cooperativity on the Free Energy Landscape of Protein Folding.Biochemistry. 2015 Jun 9;54(22):3431-41. doi: 10.1021/acs.biochem.5b00247. Epub 2015 May 28. Biochemistry. 2015. PMID: 25984766
-
Towards a consistent modeling of protein thermodynamic and kinetic cooperativity: how applicable is the transition state picture to folding and unfolding?J Mol Biol. 2002 Jan 25;315(4):899-909. doi: 10.1006/jmbi.2001.5266. J Mol Biol. 2002. PMID: 11812156
-
Molecular basis of cooperativity in protein folding. IV. CORE: a general cooperative folding model.Proteins. 1993 Oct;17(2):111-23. doi: 10.1002/prot.340170202. Proteins. 1993. PMID: 8265560
-
Limited cooperativity in protein folding.Curr Opin Struct Biol. 2016 Feb;36:58-66. doi: 10.1016/j.sbi.2015.12.001. Epub 2016 Feb 2. Curr Opin Struct Biol. 2016. PMID: 26845039 Review.
-
Protein folding intermediates and pathways studied by hydrogen exchange.Annu Rev Biophys Biomol Struct. 2000;29:213-38. doi: 10.1146/annurev.biophys.29.1.213. Annu Rev Biophys Biomol Struct. 2000. PMID: 10940248 Review.
Cited by
-
E. coli production of a multi-disulfide bonded SARS-CoV-2 Omicron BA.5 RBD exhibiting native-like biochemical and biophysical properties.Biophys Physicobiol. 2023 Sep 21;20(4):e200036. doi: 10.2142/biophysico.bppb-v20.0036. eCollection 2023. Biophys Physicobiol. 2023. PMID: 38344033 Free PMC article.
-
Protein G-quadruplex interactions and their effects on phase transitions and protein aggregation.Nucleic Acids Res. 2024 May 8;52(8):4702-4722. doi: 10.1093/nar/gkae229. Nucleic Acids Res. 2024. PMID: 38572746 Free PMC article.
-
Stability and Conformational Resilience of Protein Disulfide Isomerase.Biochemistry. 2019 Aug 27;58(34):3572-3584. doi: 10.1021/acs.biochem.9b00405. Epub 2019 Aug 16. Biochemistry. 2019. PMID: 31393106 Free PMC article.
-
Insight to Functional Conformation and Noncovalent Interactions of Protein-Protein Assembly Using MALDI Mass Spectrometry.Molecules. 2020 Oct 28;25(21):4979. doi: 10.3390/molecules25214979. Molecules. 2020. PMID: 33126406 Free PMC article. Review.
-
The Use of the Statistical Entropy in Some New Approaches for the Description of Biosystems.Entropy (Basel). 2022 Jan 24;24(2):172. doi: 10.3390/e24020172. Entropy (Basel). 2022. PMID: 35205467 Free PMC article.
References
-
- Jackson SE, Fersht AR (1991) Folding of chymotrypsin inhibitor 2. 1. Evidence for a two‐state transition. Biochemistry 30:10428–10435. - PubMed
-
- Jackson SE (1998) How do small single‐domain proteins fold? Fold Des 3:R81–R91. - PubMed
-
- Fersht A (1999) Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. Macmillan: New York.
-
- Matouschek A, Kellis JT, Serrano L, Fersht AR (1989) Mapping the transition state and pathway of protein folding by protein engineering. Nature 340:122–126. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

