High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat
- PMID: 27522358
- PMCID: PMC5069325
- DOI: 10.1007/s00122-016-2762-7
High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat
Abstract
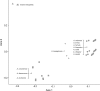
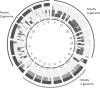
Genome analysis of 27 oat species identifies ancestral groups, delineates the D genome, and identifies ancestral origin of 21 mapped chromosomes in hexaploid oat. We investigated genomic relationships among 27 species of the genus Avena using high-density genetic markers revealed by genotyping-by-sequencing (GBS). Two methods of GBS analysis were used: one based on tag-level haplotypes that were previously mapped in cultivated hexaploid oat (A. sativa), and one intended to sample and enumerate tag-level haplotypes originating from all species under investigation. Qualitatively, both methods gave similar predictions regarding the clustering of species and shared ancestral genomes. Furthermore, results were consistent with previous phylogenies of the genus obtained with conventional approaches, supporting the robustness of whole genome GBS analysis. Evidence is presented to justify the final and definitive classification of the tetraploids A. insularis, A. maroccana (=A. magna), and A. murphyi as containing D-plus-C genomes, and not A-plus-C genomes, as is most often specified in past literature. Through electronic painting of the 21 chromosome representations in the hexaploid oat consensus map, we show how the relative frequency of matches between mapped hexaploid-derived haplotypes and AC (DC)-genome tetraploids vs. A- and C-genome diploids can accurately reveal the genome origin of all hexaploid chromosomes, including the approximate positions of inter-genome translocations. Evidence is provided that supports the continued classification of a diverged B genome in AB tetraploids, and it is confirmed that no extant A-genome diploids, including A. canariensis, are similar enough to the D genome of tetraploid and hexaploid oat to warrant consideration as a D-genome diploid.
Conflict of interest statement
Compliance with ethical standards The authors declare no breaches of ethical standards. Funding This work was funded by the Canadian Crop Genomics Initiative of the Government of Canada as part of core Agriculture and Agri-Food Canada funding. Conflict of interest The authors declare that they have no conflict of interest.
Figures


References
-
- Aung T, Zwer P, Park R, Davies P, Sidhu P, Dundas I. Hybrids of Avenasativa with two diploid wild oats (CIav6956) and (CIav7233) resistant to crown rust. Euphytica. 2010;174:189–198. doi: 10.1007/s10681-009-0111-5. - DOI
-
- Badaeva ED, Shelukhina OY, Goryunova SV, Loskutov IG, Pukhalskiy VA. Phylogenetic relationships of tetraploid AB-genome Avena species evaluated by means of cytogenetic (C-banding and FISH) and RAPD analyses. J Bot. 2010;2010:1–13. doi: 10.1155/2010/742307. - DOI
-
- Baum BR (1977) Oats: wild and cultivated. A monograph of the genus Avena L. (Poaceae). Minister of Supply and Services, Ottawa
-
- Baum BR, Fedak G. Avena atlantica, a new diploid species of the oat genus from Morocco. Can J Bot. 1985;63:1057–1060. doi: 10.1139/b85-144. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

