Plasma protein profiling in a stage defined pancreatic cancer cohort - Implications for early diagnosis
- PMID: 27522951
- PMCID: PMC5423191
- DOI: 10.1016/j.molonc.2016.07.001
Plasma protein profiling in a stage defined pancreatic cancer cohort - Implications for early diagnosis
Abstract
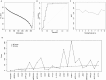
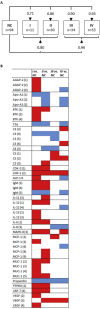
Pancreatic ductal adenocarcinoma (PDAC) is a disease where detection preceding clinical symptoms significantly increases the life expectancy of patients. In this study, a recombinant antibody microarray platform was used to analyze 213 Chinese plasma samples from PDAC patients and normal control (NC) individuals. The cohort was stratified according to disease stage, i.e. resectable disease (stage I/II), locally advanced (stage III) and metastatic disease (stage IV). Support vector machine analysis showed that all PDAC stages could be discriminated from controls and that the accuracy increased with disease progression, from stage I to IV. Patients with stage I/II PDAC could be discriminated from NC with high accuracy based on a plasma protein signature, indicating a possibility for early diagnosis and increased detection rate of surgically resectable tumors.
Keywords: Antibody microarrays; Biomarker signatures; Early detection; Pancreatic cancer; Recombinant antibodies.
Copyright © 2016 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures


References
-
- Alonzo, T.A. , Pepe, M.S. , Moskowitz, C.S. , 2002. Sample size calculations for comparative studies of medical tests for detecting presence of disease. Stat. Med. 21, 835–852. - PubMed
-
- Bellone, G. , Smirne, C. , Mauri, F.A. , Tonel, E. , Carbone, A. , Buffolino, A. , Dughera, L. , Robecchi, A. , Pirisi, M. , Emanuelli, G. , 2006. Cytokine expression profile in human pancreatic carcinoma cells and in surgical specimens: implications for survival. Cancer Immunol. Immunother. 55, 684–698. - PMC - PubMed
-
- Borrebaeck, C.A. , Wingren, C. , 2011. Recombinant antibodies for the generation of antibody arrays. Methods Mol. Biol. 785, 247–262. - PubMed
-
- Brand, R.E. , Nolen, B.M. , Zeh, H.J. , Allen, P.J. , Eloubeidi, M.A. , Goldberg, M. , Elton, E. , Arnoletti, J.P. , Christein, J.D. , Vickers, S.M. , Langmead, C.J. , Landsittel, D.P. , Whitcomb, D.C. , Grizzle, W.E. , Lokshin, A.E. , 2011. Serum biomarker panels for the detection of pancreatic cancer. Clin. Cancer Res. 17, 805–816. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

