Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells
- PMID: 27524438
- PMCID: PMC5010447
- DOI: 10.1016/j.stem.2016.07.001
Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells
Abstract
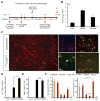
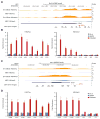
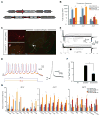
Overexpression of exogenous fate-specifying transcription factors can directly reprogram differentiated somatic cells to target cell types. Here, we show that similar reprogramming can also be achieved through the direct activation of endogenous genes using engineered CRISPR/Cas9-based transcriptional activators. We use this approach to induce activation of the endogenous Brn2, Ascl1, and Myt1l genes (BAM factors) to convert mouse embryonic fibroblasts to induced neuronal cells. This direct activation of endogenous genes rapidly remodeled the epigenetic state of the target loci and induced sustained endogenous gene expression during reprogramming. Thus, transcriptional activation and epigenetic remodeling of endogenous master transcription factors are sufficient for conversion between cell types. The rapid and sustained activation of endogenous genes in their native chromatin context by this approach may facilitate reprogramming with transient methods that avoid genomic integration and provides a new strategy for overcoming epigenetic barriers to cell fate specification.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
CONFLIC OF INTEREST C.A.G. and J.B.B. have filed patent applications related to genome engineering technologies.
Figures

Comment in
-
Advanced Technologies Lead iNto New Reprogramming Routes.Cell Stem Cell. 2016 Sep 1;19(3):286-8. doi: 10.1016/j.stem.2016.08.014. Cell Stem Cell. 2016. PMID: 27588744
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

