Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase
- PMID: 27524830
- PMCID: PMC4982548
- DOI: 10.1021/ja00134a032
Rolling-Circle RNA Synthesis: Circular Oligonucleotides as Efficient Substrates for T7 RNA Polymerase
Figures
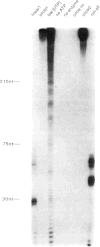
References
-
- Chamberlin MJ. In: RNA Polymerase. Losick R, Chamberlin M, editors. Cold Spring Harbor Press; Cold Spring Harbor: 1976. pp. 17–69.
- von Hippel P. H Annu Rev Biophys Biomol Struct. 1992;21:379. - PubMed
-
- Chamberlin M, Ring J. J Biol Chem. 1973;248:2235–2244. - PubMed
-
- Aiyar SE, Helmann JD, deHaseth PL. J Biol Chem. 1994;269:13179–13184. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
