Characterizing the inflammatory response in esophageal mucosal biopsies in children with eosinophilic esophagitis
- PMID: 27525061
- PMCID: PMC4973319
- DOI: 10.1038/cti.2016.30
Characterizing the inflammatory response in esophageal mucosal biopsies in children with eosinophilic esophagitis
Abstract
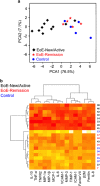
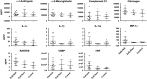
Eosinophilic esophagitis (EoE) is an emerging allergic, IgE- and non-IgE (Th2 cell)-mediated disease. There are major gaps in the understanding of the basic mechanisms that drive the persistence of EoE. We investigated whether esophageal biopsies from children with EoE demonstrate an inflammatory response that is distinct from normal controls. We prospectively enrolled 84 patients, of whom 77 were included in our analysis, aged 4-17 years (12.8±3.8 years; 81% males). Five esophageal biopsies were collected from each patient at the time of endoscopy. Intramucosal lymphocytes were isolated, phenotyped and stimulated with phorbol 12-myristate 13-acetate/ionomycin to measure their potential to produce cytokines via flow cytometry. We also performed cytokine arrays on 72-h biopsy culture supernatants. CD8(+) T cells, compared with CD4(+) T cells, synthesized more TNF-α and interferon (IFN)-γ after mitogen stimulation in the EoE-New/Active vs EoE-Remission group (P=0.0098; P=0.02) and controls (P=0.0008; P=0.03). Culture supernatants taken from explant esophageal tissue contained 13 analytes that distinguished EoE-New/Active from EoE-Remission and Controls. Principal component analysis and cluster analysis based on these analytes distinctly separated EoE-New/Active from EoE-Remission and Controls. In summary, we have identified a previously unappreciated role for CD8(+) T lymphocytes with potential to produce TNF-α and IFN-γ in EoE. Our results suggest that CD8(+) T cells have a role in the persistence or progression of EoE. We have also identified a panel of analytes produced by intact esophageal biopsies that differentiates EoE-New/Active from EoE-Remission and controls. Our results suggest that esophageal epithelial cells may have specific immune effector functions in EoE that control the type and amplitude of inflammation.
Figures




References
-
- Konikoff MR, Blanchard C, Kirby C, Buckmeier BK, Cohen MB, Heubi JE et al. Potential of blood eosinophils, eosinophil-derived neurotoxin, and eotaxin-3 as biomarkers of eosinophilic esophagitis. Clin Gastroenterol Hepatol 2006; 4: 1328–1336. - PubMed
-
- Liacouras CA, Furuta GT, Hirano I, Atkins D, Attwood SE, Bonis PA et al. Eosinophilic esophagitis: updated consensus recommendations for children and adults. J Allergy Clin Immunol 2011; 128: 3–20. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
