A systematic view on influenza induced host shutoff
- PMID: 27525483
- PMCID: PMC5028189
- DOI: 10.7554/eLife.18311
A systematic view on influenza induced host shutoff
Abstract
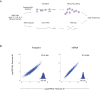
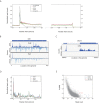
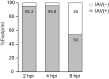
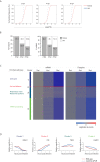
Host shutoff is a common strategy used by viruses to repress cellular mRNA translation and concomitantly allow the efficient translation of viral mRNAs. Here we use RNA-sequencing and ribosome profiling to explore the mechanisms that are being utilized by the Influenza A virus (IAV) to induce host shutoff. We show that viral transcripts are not preferentially translated and instead the decline in cellular protein synthesis is mediated by viral takeover on the mRNA pool. Our measurements also uncover strong variability in the levels of cellular transcripts reduction, revealing that short transcripts are less affected by IAV. Interestingly, these mRNAs that are refractory to IAV infection are enriched in cell maintenance processes such as oxidative phosphorylation. Furthermore, we show that the continuous oxidative phosphorylation activity is important for viral propagation. Our results advance our understanding of IAV-induced shutoff, and suggest a mechanism that facilitates the translation of genes with important housekeeping functions.
Keywords: computational biology; host shutoff; human; infectious disease; microbiology; systems biology; translation; virus infection.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

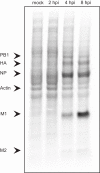

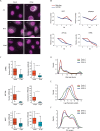

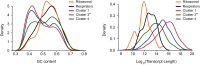
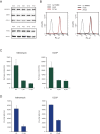
Comment in
-
Closing in on the causes of host shutoff.Elife. 2016 Sep 19;5:e20755. doi: 10.7554/eLife.20755. Elife. 2016. PMID: 27644108 Free PMC article.
References
-
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathogens. 2014;10:e18311. doi: 10.1371/journal.ppat.1003847. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

