SEDS proteins are a widespread family of bacterial cell wall polymerases
- PMID: 27525505
- PMCID: PMC5161649
- DOI: 10.1038/nature19331
SEDS proteins are a widespread family of bacterial cell wall polymerases
Abstract
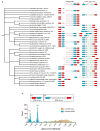
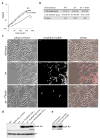
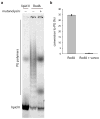
Elongation of rod-shaped bacteria is mediated by a dynamic peptidoglycan-synthetizing machinery called the Rod complex. Here we report that, in Bacillus subtilis, this complex is functional in the absence of all known peptidoglycan polymerases. Cells lacking these enzymes survive by inducing an envelope stress response that increases the expression of RodA, a widely conserved core component of the Rod complex. RodA is a member of the SEDS (shape, elongation, division and sporulation) family of proteins, which have essential but ill-defined roles in cell wall biogenesis during growth, division and sporulation. Our genetic and biochemical analyses indicate that SEDS proteins constitute a family of peptidoglycan polymerases. Thus, B. subtilis and probably most bacteria use two distinct classes of polymerase to synthesize their exoskeleton. Our findings indicate that SEDS family proteins are core cell wall synthases of the cell elongation and division machinery, and represent attractive targets for antibiotic development.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



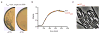

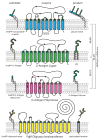


Comment in
-
Microbiology: The bacterial cell wall takes centre stage.Nature. 2016 Sep 29;537(7622):622-4. doi: 10.1038/537622a. Nature. 2016. PMID: 27680934 No abstract available.
-
Bacterial Surfaces: The Wall that SEDS Built.Curr Biol. 2016 Nov 7;26(21):R1158-R1160. doi: 10.1016/j.cub.2016.09.028. Curr Biol. 2016. PMID: 27825456
References
-
- Matteï PJ, Neves D, Dessen A. Bridging cell wall biosynthesis and bacterial morphogenesis. Current Opinion in Structural Biology. 2010;20:749–755. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

