The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan
- PMID: 27525555
- PMCID: PMC5161578
- DOI: 10.1038/nature19346
The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan
Abstract
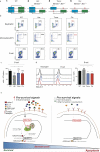
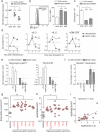
Neutrophils, eosinophils and 'classical' monocytes collectively account for about 70% of human blood leukocytes and are among the shortest-lived cells in the body. Precise regulation of the lifespan of these myeloid cells is critical to maintain protective immune responses and minimize the deleterious consequences of prolonged inflammation. However, how the lifespan of these cells is strictly controlled remains largely unknown. Here we identify a long non-coding RNA that we termed Morrbid, which tightly controls the survival of neutrophils, eosinophils and classical monocytes in response to pro-survival cytokines in mice. To control the lifespan of these cells, Morrbid regulates the transcription of the neighbouring pro-apoptotic gene, Bcl2l11 (also known as Bim), by promoting the enrichment of the PRC2 complex at the Bcl2l11 promoter to maintain this gene in a poised state. Notably, Morrbid regulates this process in cis, enabling allele-specific control of Bcl2l11 transcription. Thus, in these highly inflammatory cells, changes in Morrbid levels provide a locus-specific regulatory mechanism that allows rapid control of apoptosis in response to extracellular pro-survival signals. As MORRBID is present in humans and dysregulated in individuals with hypereosinophilic syndrome, this long non-coding RNA may represent a potential therapeutic target for inflammatory disorders characterized by aberrant short-lived myeloid cell lifespan.
Figures




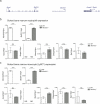

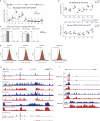
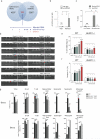
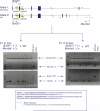



Comment in
-
Innate immune cells: A Morrbid way of stayin' alive.Nat Rev Immunol. 2016 Oct;16(10):595. doi: 10.1038/nri.2016.104. Epub 2016 Sep 12. Nat Rev Immunol. 2016. PMID: 27616588 No abstract available.
References
-
- Manz MG, Boettcher S. Emergency granulopoiesis. Nat. Rev. Immunol. 2014;14:302–314. - PubMed
-
- Ginhoux F, Jung S. Monocytes and macrophages: developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. - PubMed
-
- Heo JB, Sung S. Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science. 2011;331:76–79. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

