Comparative Single-Cell Analysis of Different E. coli Expression Systems during Microfluidic Cultivation
- PMID: 27525986
- PMCID: PMC4985164
- DOI: 10.1371/journal.pone.0160711
Comparative Single-Cell Analysis of Different E. coli Expression Systems during Microfluidic Cultivation
Abstract
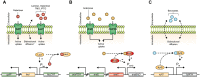
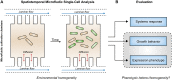
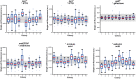
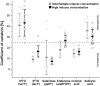
Recombinant protein production is mostly realized with large-scale cultivations and monitored at the level of the entire population. Detailed knowledge of cell-to-cell variations with respect to cellular growth and product formation is limited, even though phenotypic heterogeneity may distinctly hamper overall production yields, especially for toxic or difficult-to-express proteins. Unraveling phenotypic heterogeneity is thus a key aspect in understanding and optimizing recombinant protein production in biotechnology and synthetic biology. Here, microfluidic single-cell analysis serves as the method of choice to investigate and unmask population heterogeneities in a dynamic and spatiotemporal fashion. In this study, we report on comparative microfluidic single-cell analyses of commonly used E. coli expression systems to uncover system-inherent specifications in the synthetic M9CA growth medium. To this end, the PT7lac/LacI, the PBAD/AraC and the Pm/XylS system were systematically analyzed in order to gain detailed insights into variations of growth behavior and expression phenotypes and thus to uncover individual strengths and deficiencies at the single-cell level. Specifically, we evaluated the impact of different system-specific inducers, inducer concentrations as well as genetic modifications that affect inducer-uptake and regulation of target gene expression on responsiveness and phenotypic heterogeneity. Interestingly, the most frequently applied expression system based on E. coli strain BL21(DE3) clearly fell behind with respect to expression homogeneity and robustness of growth. Moreover, both the choice of inducer and the presence of inducer uptake systems proved crucial for phenotypic heterogeneity. Conclusively, microfluidic evaluation of different inducible E. coli expression systems and setups identified the modified lacY-deficient PT7lac/LacI as well as the Pm/XylS system with conventional m-toluic acid induction as key players for precise and robust triggering of bacterial gene expression in E. coli in a homogeneous fashion.
Conflict of interest statement
Figures


References
-
- Terpe K. Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl Microbiol Biotechnol. 2006;72: 211–22. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

