miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells
- PMID: 27526785
- PMCID: PMC4990699
- DOI: 10.1038/ncomms12436
miR-625-3p regulates oxaliplatin resistance by targeting MAP2K6-p38 signalling in human colorectal adenocarcinoma cells
Abstract
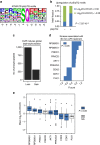
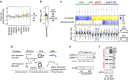
Oxaliplatin resistance in colorectal cancers (CRC) is a major medical problem, and predictive markers are urgently needed. Recently, miR-625-3p was reported as a promising predictive marker. Herein, we show that miR-625-3p functionally induces oxaliplatin resistance in CRC cells, and identify the signalling networks affected by miR-625-3p. We show that the p38 MAPK activator MAP2K6 is a direct target of miR-625-3p, and, accordingly, is downregulated in non-responder patients of oxaliplatin therapy. miR-625-3p-mediated resistance is reversed by anti-miR-625-3p treatment and ectopic expression of a miR-625-3p insensitive MAP2K6 variant. In addition, reduction of p38 signalling by using siRNAs, chemical inhibitors or expression of a dominant-negative MAP2K6 protein induces resistance to oxaliplatin. Transcriptome, proteome and phosphoproteome profiles confirm inactivation of MAP2K6-p38 signalling as one likely mechanism of oxaliplatin resistance. Our study shows that miR-625-3p induces oxaliplatin resistance by abrogating MAP2K6-p38-regulated apoptosis and cell cycle control networks, and corroborates the predictive power of miR-625-3p.
Figures
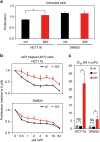
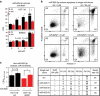

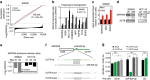

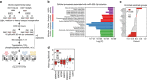

References
-
- Jemal A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011). - PubMed
-
- Schmoll H. J. et al. ESMO Consensus Guidelines for management of patients with colon and rectal cancer: a personalized approach to clinical decision making. Ann. Oncol. 23, 2479–2516 (2012). - PubMed
-
- Bartel D. P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004). - PubMed
-
- Blower P. E. et al. MicroRNAs modulate the chemosensitivity of tumor cells. Mol. Cancer Ther. 7, 1–9 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

