Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis
- PMID: 27527070
- PMCID: PMC4986186
- DOI: 10.1186/s12864-016-2965-y
Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis
Abstract
Background: An estimated 15,000 children and adolescents under the age of 19 years are diagnosed with leukemia, lymphoma and other tumors in the USA every year. All children and adolescent acute leukemia patients will undergo chemotherapy as part of their treatment regimen. Fortunately, survival rates for most pediatric cancers have improved at a remarkable pace over the past three decades, and the overall survival rate is greater than 90 % today. However, significant differences in survival rate have been found in different age groups (94 % in 1-9.99 years, 82 % in ≥10 years and 76 % in ≥15 years). ALL accounts for about three out of four cases of childhood leukemia. Intensive chemotherapy treatment coupled with prophylactic or therapeutic antibiotic use could potentially have a long-term effect on the resident gastrointestinal (GI) microbiome. The composition of GI microbiome and its changes upon chemotherapy in pediatric and adolescent leukemia patients is poorly understood. In this study, using 16S rRNA marker gene sequences we profile the GI microbial communities of pediatric and adolescent acute leukemia patients before and after chemotherapy treatment and compare with the microbiota of their healthy siblings.
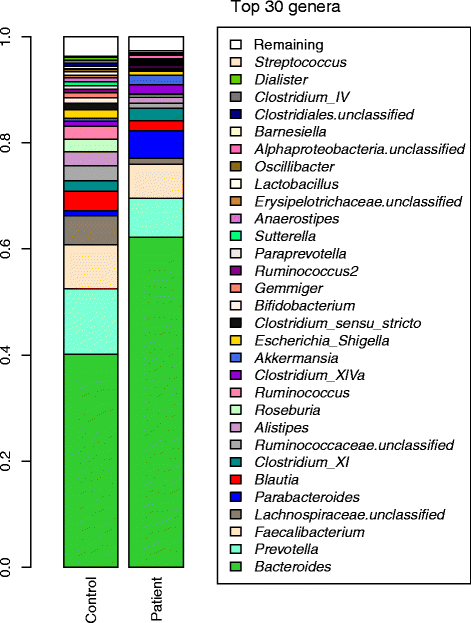
Results: Our study cohort consisted of 51 participants, made up of matched pediatric and adolescent patients with ALL and a healthy sibling. We elucidated and compared the GI microbiota profiles of patients and their healthy sibling controls via analysis of 16S rRNA gene sequencing data. We assessed the GI microbiota composition in pediatric and adolescent patients with ALL during the course of chemotherapy by comparing stool samples taken before chemotherapy with stool samples collected at varying time points during the chemotherapeutic treatment. The microbiota profiles of both patients and control sibling groups are dominated by members of Bacteroides, Prevotella, and Faecalibacterium. At the genus level, both groups share many taxa in common, but the microbiota diversity of the patient group is significantly lower than that of the control group. It was possible to distinguish between the patient and control groups based on their microbiota profiles. The top taxa include Anaerostipes, Coprococcus, Roseburia, and Ruminococcus2 with relatively higher abundance in the control group. The observed microbiota changes are likely the result of several factors including a direct influence of therapeutic compounds on the gut flora and an indirect effect of chemotherapy on the immune system, which, in turn, affects the microbiota.
Conclusions: This study provides significant information on GI microbiota populations in immunocompromised children and opens up the potential for developing novel diagnostics based on stool tests and therapies to improve the dysbiotic condition of the microbiota at the time of diagnosis and in the earliest stages of chemotherapy.
Keywords: 16S rRNA gene sequencing; Gastrointestinal microbiota; Pediatric leukemia; Ribosomal RNA; rRNA.
Figures



References
-
- Bates D, Maechler M, Bolker B, Walker S. R package version 02–3. 2014. lme4: Linear mixed-effects models using Eigen and S4. R package version 1.1-5.
-
- Holler E, Butzhammer P, Schmid K, Hundsrucker C, Koestler J, Peter K, Zhu W, Sporrer D, Hehlgans T, Kreutz M, et al. Metagenomic analysis of the stool microbiome in patients receiving allogeneic stem cell transplantation: loss of diversity is associated with use of systemic antibiotics and more pronounced in gastrointestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2014;20(5):640–645. doi: 10.1016/j.bbmt.2014.01.030. - DOI - PMC - PubMed
-
- Hudson MM, Mertens AC, Yasui Y, Hobbie W, Chen H, Gurney JG, Yeazel M, Recklitis CJ, Marina N, Robison LR, et al. Health status of adult long-term survivors of childhood cancer: a report from the Childhood Cancer Survivor Study. JAMA. 2003;290(12):1583–1592. doi: 10.1001/jama.290.12.1583. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

