Resistance to Macrolide Antibiotics in Public Health Pathogens
- PMID: 27527699
- PMCID: PMC5046686
- DOI: 10.1101/cshperspect.a025395
Resistance to Macrolide Antibiotics in Public Health Pathogens
Abstract
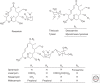
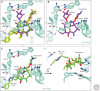
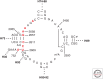
Macrolide resistance mechanisms can be target-based with a change in a 23S ribosomal RNA (rRNA) residue or a mutation in ribosomal protein L4 or L22 affecting the ribosome's interaction with the antibiotic. Alternatively, mono- or dimethylation of A2058 in domain V of the 23S rRNA by an acquired rRNA methyltransferase, the product of an erm (erythromycin ribosome methylation) gene, can interfere with antibiotic binding. Acquired genes encoding efflux pumps, most predominantly mef(A) + msr(D) in pneumococci/streptococci and msr(A/B) in staphylococci, also mediate resistance. Drug-inactivating mechanisms include phosphorylation of the 2'-hydroxyl of the amino sugar found at position C5 by phosphotransferases and hydrolysis of the macrocyclic lactone by esterases. These acquired genes are regulated by either translation or transcription attenuation, largely because cells are less fit when these genes, especially the rRNA methyltransferases, are highly induced or constitutively expressed. The induction of gene expression is cleverly tied to the mechanism of action of macrolides, relying on antibiotic-bound ribosomes stalled at specific sequences of nascent polypeptides to promote transcription or translation of downstream sequences.
Copyright © 2016 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures

References
-
- Abbassi MS, Torres C, Achour W, Vinue L, Saenz Y, Costa D, Bouchami O, Ben Hassen A. 2008. Genetic characterisation of CTX-M-15-producing Klebsiella pneumoniae and Escherichia coli strains isolated from stem cell transplant patients in Tunisia. Int J Antimicrob Agents 32: 308–314. - PubMed
-
- Amezaga MR, McKenzie H. 2006. Molecular epidemiology of macrolide resistance in β-haemolytic streptococci of Lancefield groups A, B, C and G and evidence for a new mef element in group G streptococci that carries allelic variants of mef and msr(D). J Antimicrob Chemother 57: 443–449. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous
