Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens
- PMID: 27527800
- PMCID: PMC4990709
- DOI: 10.1038/ncomms12506
Human TRAV1-2-negative MR1-restricted T cells detect S. pyogenes and alternatives to MAIT riboflavin-based antigens
Abstract
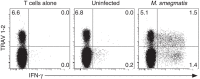
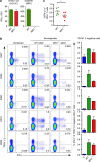
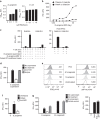
Mucosal-associated invariant T (MAIT) cells are thought to detect microbial antigens presented by the HLA-Ib molecule MR1 through the exclusive use of a TRAV1-2-containing TCRα. Here we use MR1 tetramer staining and ex vivo analysis with mycobacteria-infected MR1-deficient cells to demonstrate the presence of functional human MR1-restricted T cells that lack TRAV1-2. We characterize an MR1-restricted clone that expresses the TRAV12-2 TCRα, which lacks residues previously shown to be critical for MR1-antigen recognition. In contrast to TRAV1-2(+) MAIT cells, this TRAV12-2-expressing clone displays a distinct pattern of microbial recognition by detecting infection with the riboflavin auxotroph Streptococcus pyogenes. As known MAIT antigens are derived from riboflavin metabolites, this suggests that TRAV12-2(+) clone recognizes unique antigens. Thus, MR1-restricted T cells can discriminate between microbes in a TCR-dependent manner. We postulate that additional MR1-restricted T-cell subsets may play a unique role in defence against infection by broadening the recognition of microbial metabolites.
Figures



Similar articles
-
MAIT, MR1, microbes and riboflavin: a paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules?Immunogenetics. 2016 Aug;68(8):537-48. doi: 10.1007/s00251-016-0927-9. Epub 2016 Jul 8. Immunogenetics. 2016. PMID: 27393664 Review.
-
MHC class I-related molecule, MR1, and mucosal-associated invariant T cells.Immunol Rev. 2016 Jul;272(1):120-38. doi: 10.1111/imr.12423. Immunol Rev. 2016. PMID: 27319347 Review.
-
MAIT cells and MR1-antigen recognition.Curr Opin Immunol. 2017 Jun;46:66-74. doi: 10.1016/j.coi.2017.04.002. Epub 2017 May 8. Curr Opin Immunol. 2017. PMID: 28494326 Review.
-
Antigen Recognition by MR1-Reactive T Cells; MAIT Cells, Metabolites, and Remaining Mysteries.Front Immunol. 2020 Aug 27;11:1961. doi: 10.3389/fimmu.2020.01961. eCollection 2020. Front Immunol. 2020. PMID: 32973800 Free PMC article. Review.
-
Diverse MR1-restricted T cells in mice and humans.Nat Commun. 2019 May 21;10(1):2243. doi: 10.1038/s41467-019-10198-w. Nat Commun. 2019. PMID: 31113973 Free PMC article.
Cited by
-
Characterization of the TCRβ repertoire of peripheral MR1-restricted MAIT cells in psoriasis vulgaris patients.Sci Rep. 2023 Nov 28;13(1):20990. doi: 10.1038/s41598-023-48321-z. Sci Rep. 2023. PMID: 38017021 Free PMC article.
-
Generation of MR1-Restricted T Cell Clones by Limiting Dilution Cloning of MR1 Tetramer+ Cells.Methods Mol Biol. 2020;2098:219-235. doi: 10.1007/978-1-0716-0207-2_15. Methods Mol Biol. 2020. PMID: 31792826 Free PMC article.
-
MR1 recycling and blockade of endosomal trafficking reveal distinguishable antigen presentation pathways between Mycobacterium tuberculosis infection and exogenously delivered antigens.Sci Rep. 2019 Mar 18;9(1):4797. doi: 10.1038/s41598-019-41402-y. Sci Rep. 2019. PMID: 30886396 Free PMC article.
-
MR1-restricted T cell clonotypes are associated with "resistance" to Mycobacterium tuberculosis infection.JCI Insight. 2024 May 8;9(9):e166505. doi: 10.1172/jci.insight.166505. JCI Insight. 2024. PMID: 38716731 Free PMC article.
-
Translating Unconventional T Cells and Their Roles in Leukemia Antitumor Immunity.J Immunol Res. 2021 Jan 7;2021:6633824. doi: 10.1155/2021/6633824. eCollection 2021. J Immunol Res. 2021. PMID: 33506055 Free PMC article. Review.
References
-
- Le Bourhis L. et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 11, 701–708 (2010). - PubMed
-
- Corbett A. J. et al. T-cell activation by transitory neo-antigens derived from distinct microbial pathways. Nature 509, 361–365 (2014). - PubMed
-
- Kjer-Nielsen L. et al. MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491, 717–723 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

