Fluorescent reporter analysis revealed the timing and localization of AVR-Pia expression, an avirulence effector of Magnaporthe oryzae
- PMID: 27528510
- PMCID: PMC6638300
- DOI: 10.1111/mpp.12468
Fluorescent reporter analysis revealed the timing and localization of AVR-Pia expression, an avirulence effector of Magnaporthe oryzae
Abstract
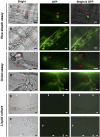
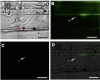
In order to facilitate infection, the rice blast pathogen Magnaporthe oryzae secretes an abundance of proteins, including avirulence effectors, to diminish its host's defences. Avirulence effectors are recognized by host resistance proteins and trigger the host's hypersensitive response, which is a rapid and effective form of innate plant immunity. An understanding of the underlying molecular mechanisms of such interactions is crucial for the development of strategies to control disease. However, the expression and secretion of certain effector proteins, such as AVR-Pia, have yet to be reported. Reverse transcription-polymerase chain reaction (RT-PCR) revealed that AVR-Pia was only expressed during infection. Fluorescently labelled AVR-Pia indicated that AVR-Pia expression was induced during appressorial differentiation in the cells of both rice and onion, as well as in a penetration-deficient (Δpls1) mutant capable of developing melanized appressoria, but unable to penetrate host cells, suggesting that AVR-Pia expression is independent of fungal penetration. Using live-cell imaging, we also documented the co-localization of green fluorescent protein (GFP)-labelled AVR-Pia and monomeric red fluorescent protein (mRFP)-labelled PWL2, which indicates that AVR-Pia accumulates in biotrophic interfacial complexes before being delivered to the plant cytosol. Together, these results suggest that AVR-Pia is a cytoplasmic effector that is expressed at the onset of appressorial differentiation and is translocated to the biotrophic interfacial complex, and then into the host's cytoplasm.
Keywords: AVR-Pia; BIC, Pyricularia oryzae; appressorium; avirulence effector; rice.
© 2016 BSPP AND JOHN WILEY & SONS LTD.
Figures





Similar articles
-
The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery.Plant J. 2013 Apr;74(1):1-12. doi: 10.1111/tpj.12099. Epub 2013 Mar 4. Plant J. 2013. PMID: 23279638
-
Recent advances in rice blast effector research.Curr Opin Plant Biol. 2010 Aug;13(4):434-41. doi: 10.1016/j.pbi.2010.04.012. Epub 2010 Jun 2. Curr Opin Plant Biol. 2010. PMID: 20627803 Review.
-
Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae.Plant Cell. 2009 May;21(5):1573-91. doi: 10.1105/tpc.109.066324. Epub 2009 May 19. Plant Cell. 2009. PMID: 19454732 Free PMC article.
-
Visualizing the Movement of Magnaporthe oryzae Effector Proteins in Rice Cells During Infection.Methods Mol Biol. 2018;1848:103-117. doi: 10.1007/978-1-4939-8724-5_9. Methods Mol Biol. 2018. PMID: 30182232
-
Effector-triggered susceptibility by the rice blast fungus Magnaporthe oryzae.New Phytol. 2024 Feb;241(3):1007-1020. doi: 10.1111/nph.19446. Epub 2023 Dec 10. New Phytol. 2024. PMID: 38073141 Review.
Cited by
-
Genome sequence and spore germination-associated transcriptome analysis of Corynespora cassiicola from cucumber.BMC Microbiol. 2020 Jul 8;20(1):199. doi: 10.1186/s12866-020-01873-w. BMC Microbiol. 2020. PMID: 32641051 Free PMC article.
-
The essential effector SCRE1 in Ustilaginoidea virens suppresses rice immunity via a small peptide region.Mol Plant Pathol. 2020 Apr;21(4):445-459. doi: 10.1111/mpp.12894. Epub 2020 Feb 22. Mol Plant Pathol. 2020. PMID: 32087618 Free PMC article.
-
The roles of Magnaporthe oryzae avirulence effectors involved in blast resistance/susceptibility.Front Plant Sci. 2024 Oct 9;15:1478159. doi: 10.3389/fpls.2024.1478159. eCollection 2024. Front Plant Sci. 2024. PMID: 39445147 Free PMC article. Review.
-
Recent Advances in Effector Research of Magnaporthe oryzae.Biomolecules. 2023 Nov 14;13(11):1650. doi: 10.3390/biom13111650. Biomolecules. 2023. PMID: 38002332 Free PMC article. Review.
-
A nonclassically secreted effector of Magnaporthe oryzae targets host nuclei and plays important roles in fungal growth and plant infection.Mol Plant Pathol. 2023 Sep;24(9):1093-1106. doi: 10.1111/mpp.13356. Epub 2023 Jun 12. Mol Plant Pathol. 2023. PMID: 37306516 Free PMC article.
References
-
- Abe, A. , Elegado, E. and Sone, T. (2006) Construction of pDESTR, a GATEWAY vector for gene disruption in filamentous fungi. Curr. Microbiol. 52, 210–215. - PubMed
-
- Ballini, E. , Morel, J.B. , Droc, G. , Price, A. , Courtois, B. , Notteghem, J.L. and Tharreau, D. (2008) A genome‐wide meta‐analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol. Plant–Microbe Interact. 21, 859–868. - PubMed
-
- Clergeot, P.H. , Gourgues, M. , Cots, J. , Laurans, F. , Latorse, M.P. , Pepin, R. , Tharreau, D. , Notteghem, J.L. and Lebrun, M.H. (2001) PLS1, a gene encoding a tetraspanin‐like protein, is required for penetration of rice leaf by the fungal pathogen Magnaporthe grisea . Proc. Natl. Acad. Sci. USA, 98, 6963–6968. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

