The Evolutionarily-conserved Polyadenosine RNA Binding Protein, Nab2, Cooperates with Splicing Machinery to Regulate the Fate of pre-mRNA
- PMID: 27528618
- PMCID: PMC5064217
- DOI: 10.1128/MCB.00402-16
The Evolutionarily-conserved Polyadenosine RNA Binding Protein, Nab2, Cooperates with Splicing Machinery to Regulate the Fate of pre-mRNA
Abstract
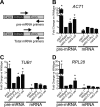
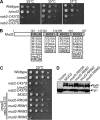
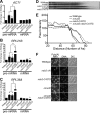
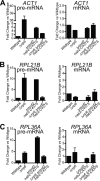
Numerous RNA binding proteins are deposited onto an mRNA transcript to modulate post-transcriptional processing events ensuring proper mRNA maturation. Defining the interplay between RNA binding proteins that couple mRNA biogenesis events is crucial for understanding how gene expression is regulated. To explore how RNA binding proteins control mRNA processing, we investigated a role for the evolutionarily conserved polyadenosine RNA binding protein, Nab2, in mRNA maturation within the nucleus. This work reveals that nab2 mutant cells accumulate intron-containing pre-mRNA in vivo We extend this analysis to identify genetic interactions between mutant alleles of nab2 and genes encoding the splicing factor, MUD2, and the RNA exosome, RRP6, with in vivo consequences of altered pre-mRNA splicing and poly(A) tail length control. As further evidence linking Nab2 proteins to splicing, an unbiased proteomic analysis of vertebrate Nab2, ZC3H14, identifies physical interactions with numerous components of the spliceosome. We validated the interaction between ZC3H14 and U2AF2/U2AF65 Taking all the findings into consideration, we present a model where Nab2/ZC3H14 interacts with spliceosome components to allow proper coupling of splicing with subsequent mRNA processing steps contributing to a kinetic proofreading step that allows properly processed mRNA to exit the nucleus and escape Rrp6-dependent degradation.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures






References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
