Epigenetic Mechanisms in Bone Biology and Osteoporosis: Can They Drive Therapeutic Choices?
- PMID: 27529237
- PMCID: PMC5000726
- DOI: 10.3390/ijms17081329
Epigenetic Mechanisms in Bone Biology and Osteoporosis: Can They Drive Therapeutic Choices?
Abstract
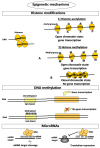
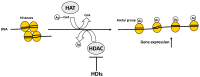
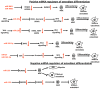
Osteoporosis is a complex multifactorial disorder of the skeleton. Genetic factors are important in determining peak bone mass and structure, as well as the predisposition to bone deterioration and fragility fractures. Nonetheless, genetic factors alone are not sufficient to explain osteoporosis development and fragility fracture occurrence. Indeed, epigenetic factors, representing a link between individual genetic aspects and environmental influences, are also strongly suspected to be involved in bone biology and osteoporosis. Recently, alterations in epigenetic mechanisms and their activity have been associated with aging. Also, bone metabolism has been demonstrated to be under the control of epigenetic mechanisms. Runt-related transcription factor 2 (RUNX2), the master transcription factor of osteoblast differentiation, has been shown to be regulated by histone deacetylases and microRNAs (miRNAs). Some miRNAs were also proven to have key roles in the regulation of Wnt signalling in osteoblastogenesis, and to be important for the positive or negative regulation of both osteoblast and osteoclast differentiation. Exogenous and environmental stimuli, influencing the functionality of epigenetic mechanisms involved in the regulation of bone metabolism, may contribute to the development of osteoporosis and other bone disorders, in synergy with genetic determinants. The progressive understanding of roles of epigenetic mechanisms in normal bone metabolism and in multifactorial bone disorders will be very helpful for a better comprehension of disease pathogenesis and translation of this information into clinical practice. A deep understanding of these mechanisms could help in the future tailoring of proper individual treatments, according to precision medicine's principles.
Keywords: DNA methylation; fragility fracture; gene expression; histone modifications; microRNAs; precision medicine.
Figures
Similar articles
-
The emerging role of microRNAs in bone remodeling and its therapeutic implications for osteoporosis.Biosci Rep. 2018 Jun 21;38(3):BSR20180453. doi: 10.1042/BSR20180453. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29848766 Free PMC article. Review.
-
The Role of Epigenomics in Osteoporosis and Osteoporotic Vertebral Fracture.Int J Mol Sci. 2020 Dec 11;21(24):9455. doi: 10.3390/ijms21249455. Int J Mol Sci. 2020. PMID: 33322579 Free PMC article. Review.
-
[Epigenetic Aspects of Osteoporosis].Vestn Ross Akad Med Nauk. 2015;(5):541-8. doi: 10.15690/vramn.v70.i5.1440. Vestn Ross Akad Med Nauk. 2015. PMID: 26846079 Review. Russian.
-
Deregulated miRNAs in bone health: Epigenetic roles in osteoporosis.Bone. 2019 May;122:52-75. doi: 10.1016/j.bone.2019.02.013. Epub 2019 Feb 14. Bone. 2019. PMID: 30772601 Review.
-
Epigenetic mechanisms in bone.Clin Chem Lab Med. 2014 May;52(5):589-608. doi: 10.1515/cclm-2013-0770. Clin Chem Lab Med. 2014. PMID: 24353145 Review.
Cited by
-
Osteogenic Commitment of Human Periodontal Ligament Cells Is Predetermined by Methylation, Chromatin Accessibility and Expression of Key Transcription Factors.Cells. 2022 Mar 26;11(7):1126. doi: 10.3390/cells11071126. Cells. 2022. PMID: 35406691 Free PMC article.
-
Role of Dietary Supplements and Probiotics in Modulating Microbiota and Bone Health: The Gut-Bone Axis.Cells. 2022 Feb 21;11(4):743. doi: 10.3390/cells11040743. Cells. 2022. PMID: 35203401 Free PMC article. Review.
-
Identification of novel genes in aging osteoblasts using next-generation sequencing and bioinformatics.Oncotarget. 2017 Nov 28;8(69):113598-113613. doi: 10.18632/oncotarget.22748. eCollection 2017 Dec 26. Oncotarget. 2017. PMID: 29371932 Free PMC article.
-
Expression Profiles of Exosomal miRNAs in Gaucher Patients and Their Association With Severity of Bone Involvement.J Inherit Metab Dis. 2025 Jul;48(4):e70061. doi: 10.1002/jimd.70061. J Inherit Metab Dis. 2025. PMID: 40619747 Free PMC article.
-
From Genomics to Metabolomics: Molecular Insights into Osteoporosis for Enhanced Diagnostic and Therapeutic Approaches.Biomedicines. 2024 Oct 18;12(10):2389. doi: 10.3390/biomedicines12102389. Biomedicines. 2024. PMID: 39457701 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

