Phylodynamic Characterization of an Ocular-Tropism Coxsackievirus A24 Variant
- PMID: 27529556
- PMCID: PMC4987047
- DOI: 10.1371/journal.pone.0160672
Phylodynamic Characterization of an Ocular-Tropism Coxsackievirus A24 Variant
Abstract
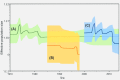
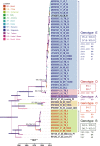
Recent phylodynamic studies have focused on using tree topology patterns to elucidate interactions among the epidemiological, evolutionary, and demographic characteristics of infectious agents. However, because studies of viral phylodynamics tend to focus on epidemic outbreaks, tree topology signatures of tissue-tropism pathogens might not be clearly identified. Therefore, this study used a novel Bayesian evolutionary approach to analyze the A24 variant of coxsackievirus (CV-A24v), an ocular-tropism agent of acute hemorrhagic conjunctivitis. Analyses of the 915-nucleotide VP1 and 690-nt 3Dpol regions of 21 strains isolated in Taiwan and worldwide during 1985-2010 revealed a clear chronological trend in both the VP1 and 3Dpol phylogenetic trees: the emergence of a single dominant cluster in each outbreak. The VP1 sequences included three genotypes: GI (prototype), GIII (isolated 1985-1999), and GIV (isolated after 2000); no VP1 sequences from GII strains have been deposited in GenBank. Another five genotypes identified in the 3Dpol region had support values >0.9. Geographic and demographic transitions among CV-A24v clusters were clearly identified by Bayes algorithm. The transmission route was mapped from India to China and then to Taiwan, and each prevalent viral population declined before new clusters emerged. Notably, the VP1 and 3Dpol genes had high nucleotide sequence similarities (94.1% and 95.2%, respectively). The lack of co-circulating lineages and narrow tissue tropism affected the CV-A24v gene pool.
Conflict of interest statement
Figures


References
-
- Lin KH, Chern CL, Chu PY, Chang CH, Wang HL, Sheu MM, et al. Genetic analysis of recent Taiwanese isolates of a variant of coxsackievirus A24. J Med Virol. 2001; 64:269–274. - PubMed
-
- Khetsuriani N, Ashley L-F, Oberst MS, Pallansch MA. Enterovirus surveillance—United States, 1970–2005. MMWR Surveill Summ 2006; 15:1–20. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

