New evidence for grain specific C4 photosynthesis in wheat
- PMID: 27530078
- PMCID: PMC4987656
- DOI: 10.1038/srep31721
New evidence for grain specific C4 photosynthesis in wheat
Abstract
The C4 photosynthetic pathway evolved to allow efficient CO2 capture by plants where effective carbon supply may be limiting as in hot or dry environments, explaining the high growth rates of C4 plants such as maize. Important crops such as wheat and rice are C3 plants resulting in efforts to engineer them to use the C4 pathway. Here we show the presence of a C4 photosynthetic pathway in the developing wheat grain that is absent in the leaves. Genes specific for C4 photosynthesis were identified in the wheat genome and found to be preferentially expressed in the photosynthetic pericarp tissue (cross- and tube-cell layers) of the wheat caryopsis. The chloroplasts exhibit dimorphism that corresponds to chloroplasts of mesophyll- and bundle sheath-cells in leaves of classical C4 plants. Breeding to optimize the relative contributions of C3 and C4 photosynthesis may adapt wheat to climate change, contributing to wheat food security.
Figures


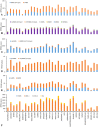
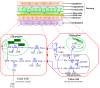
Comment in
-
Commentary: New evidence for grain specific C4 photosynthesis in wheat.Front Plant Sci. 2016 Oct 19;7:1537. doi: 10.3389/fpls.2016.01537. eCollection 2016. Front Plant Sci. 2016. PMID: 27807439 Free PMC article. No abstract available.
-
Wheat genomics: Seeds of C4 photosynthesis.Nat Plants. 2016 Nov 3;2(11):16172. doi: 10.1038/nplants.2016.172. Nat Plants. 2016. PMID: 27808227 No abstract available.
Similar articles
-
The Expression of TaRca2-α Gene Associated with Net Photosynthesis Rate, Biomass and Grain Yield in Bread Wheat (Triticum aestivum L.) under Field Conditions.PLoS One. 2016 Aug 22;11(8):e0161308. doi: 10.1371/journal.pone.0161308. eCollection 2016. PLoS One. 2016. PMID: 27548477 Free PMC article.
-
The Metabolite Pathway between Bundle Sheath and Mesophyll: Quantification of Plasmodesmata in Leaves of C3 and C4 Monocots.Plant Cell. 2016 Jun;28(6):1461-71. doi: 10.1105/tpc.16.00155. Epub 2016 Jun 10. Plant Cell. 2016. PMID: 27288224 Free PMC article.
-
Exaptation of ancestral cell-identity networks enables C4 photosynthesis.Nature. 2024 Dec;636(8041):143-150. doi: 10.1038/s41586-024-08204-3. Epub 2024 Nov 20. Nature. 2024. PMID: 39567684 Free PMC article.
-
Starch Accumulation in the Bundle Sheaths of C3 Plants: A Possible Pre-Condition for C4 Photosynthesis.Plant Cell Physiol. 2016 May;57(5):890-6. doi: 10.1093/pcp/pcw046. Epub 2016 Mar 2. Plant Cell Physiol. 2016. PMID: 26936788 Review.
-
[C4 type photosynthesis].Postepy Biochem. 2012;58(1):44-53. Postepy Biochem. 2012. PMID: 23214128 Review. Polish.
Cited by
-
Using Biotechnology-Led Approaches to Uplift Cereal and Food Legume Yields in Dryland Environments.Front Plant Sci. 2018 Aug 27;9:1249. doi: 10.3389/fpls.2018.01249. eCollection 2018. Front Plant Sci. 2018. PMID: 30210519 Free PMC article. Review.
-
Exploring natural variation of photosynthesis in a site-specific manner: evolution, progress, and prospects.Planta. 2019 Oct;250(4):1033-1050. doi: 10.1007/s00425-019-03223-1. Epub 2019 Jun 28. Planta. 2019. PMID: 31254100 Review.
-
Evolution of an intermediate C4 photosynthesis in the non-foliar tissues of the Poaceae.Photosynth Res. 2022 Sep;153(3):125-134. doi: 10.1007/s11120-022-00926-7. Epub 2022 Jun 1. Photosynth Res. 2022. PMID: 35648247 Review.
-
Photosynthetic contribution and characteristics of cucumber stems and petioles.BMC Plant Biol. 2021 Oct 6;21(1):454. doi: 10.1186/s12870-021-03233-w. BMC Plant Biol. 2021. PMID: 34615487 Free PMC article.
-
Understanding grain development in the Poaceae family by comparing conserved and distinctive pathways through omics studies in wheat and maize.Front Plant Sci. 2024 Jul 18;15:1393140. doi: 10.3389/fpls.2024.1393140. eCollection 2024. Front Plant Sci. 2024. PMID: 39100085 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

