Gene content dissimilarity for subclassification of highly similar microbial strains
- PMID: 27530250
- PMCID: PMC4988056
- DOI: 10.1186/s12864-016-2991-9
Gene content dissimilarity for subclassification of highly similar microbial strains
Abstract
Background: Identification and classification of highly similar microbial strains is a challenging issue in microbiology, ecology and evolutionary biology. Among various available approaches, gene content analysis is also at the core of microbial taxonomy. However, no threshold has been determined for grouping microorgnisms to different taxonomic levels, and it is still not clear that to what extent genomic fluidity should occur to form a microbial taxonomic group.
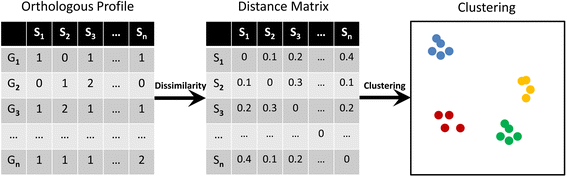
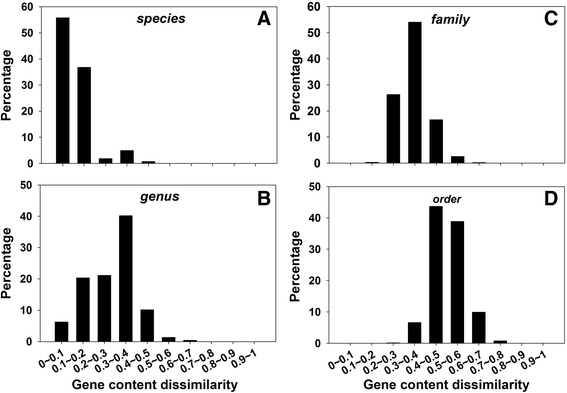
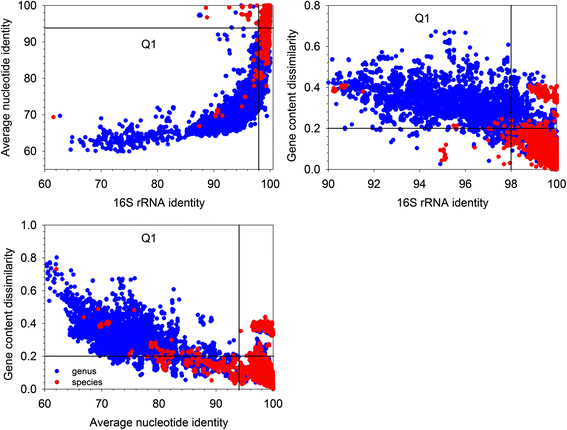
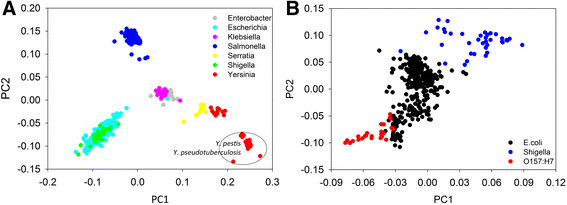
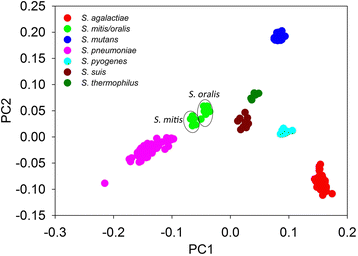
Results: By taking advantage of the eggNOG database for orthologous groups, we calculated gene content dissimilarity among different microbial strains based on the orthologous gene profiles and tested the possibility of applying gene content dissimilarity as a quantitative index in classifying microbial taxonomic groups, as well as its potential application in subclassification of highly similar microbial strains. Evaluation of gene content dissimilarity to completed microbial genomes at different taxonomic levels suggested that cutoffs of 0.2 and 0.4 can be respectively used for species and family delineation, and that 0.2 gene content dissimilarity cutoff approximately corresponded to 98 % 16S rRNA gene identity and 94 % ANI for microbial species delineation. Furthermore, application of gene content dissimilarity to highly similar microbial strains suggested it as an effective approach in classifying closely related microorganisms into subgroups.
Conclusions: This approach is especially useful in identifying pathogens from commensals in clinical microbiology. It also provides novel insights into how genomic fluidity is linked with microbial taxonomy.
Keywords: Gene content dissimilarity; Genomic fluidity; Highly similar strains; Microbial subclassification.
Figures
Similar articles
-
CAMITAX: Taxon labels for microbial genomes.Gigascience. 2020 Jan 1;9(1):giz154. doi: 10.1093/gigascience/giz154. Gigascience. 2020. PMID: 31909794 Free PMC article.
-
Computational integration of genomic traits into 16S rDNA microbiota sequencing studies.Gene. 2014 Oct 1;549(1):186-91. doi: 10.1016/j.gene.2014.07.066. Epub 2014 Jul 30. Gene. 2014. PMID: 25084126
-
A comparison of methods for clustering 16S rRNA sequences into OTUs.PLoS One. 2013 Aug 13;8(8):e70837. doi: 10.1371/journal.pone.0070837. eCollection 2013. PLoS One. 2013. PMID: 23967117 Free PMC article.
-
Recovering complete and draft population genomes from metagenome datasets.Microbiome. 2016 Mar 8;4:8. doi: 10.1186/s40168-016-0154-5. Microbiome. 2016. PMID: 26951112 Free PMC article. Review.
-
[A review on the bioinformatics pipelines for metagenomic research].Dongwuxue Yanjiu. 2012 Dec;33(6):574-85. doi: 10.3724/SP.J.1141.2012.06574. Dongwuxue Yanjiu. 2012. PMID: 23266976 Review. Chinese.
Cited by
-
Discovery of novel treponemes associated with pododermatitis in elk (Cervus canadensis).Appl Environ Microbiol. 2024 Jun 18;90(6):e0010524. doi: 10.1128/aem.00105-24. Epub 2024 May 14. Appl Environ Microbiol. 2024. PMID: 38742897 Free PMC article.
-
On a Non-Discrete Concept of Prokaryotic Species.Microorganisms. 2020 Nov 4;8(11):1723. doi: 10.3390/microorganisms8111723. Microorganisms. 2020. PMID: 33158054 Free PMC article.
-
Mangrovimonas cancribranchiae sp. nov., a novel bacterial species associated with the gills of the fiddler crab Cranuca inversa (Brachyura, Ocypodidae) from Red Sea mangroves.Int J Syst Evol Microbiol. 2024 Jun;74(6):006415. doi: 10.1099/ijsem.0.006415. Int J Syst Evol Microbiol. 2024. PMID: 38865172 Free PMC article.
-
GET_PHYLOMARKERS, a Software Package to Select Optimal Orthologous Clusters for Phylogenomics and Inferring Pan-Genome Phylogenies, Used for a Critical Geno-Taxonomic Revision of the Genus Stenotrophomonas.Front Microbiol. 2018 May 1;9:771. doi: 10.3389/fmicb.2018.00771. eCollection 2018. Front Microbiol. 2018. PMID: 29765358 Free PMC article.
-
Diverse microbiome functions, limited temporal variation and substantial genomic conservation within sedimentary and granite rock deep underground research laboratories.Environ Microbiome. 2024 Dec 18;19(1):105. doi: 10.1186/s40793-024-00649-3. Environ Microbiome. 2024. PMID: 39696556 Free PMC article.
References
-
- Rodriguez-R LM, Konstantinidis KT. Bypassing cultivation to identify bacterial species. Microbe. 2014;9(3):111–8.
-
- Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6(6):431–40. - PubMed
-
- STACKEBRANDT E, GOEBEL BM. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol. 1994;44(4):846–9. doi: 10.1099/00207713-44-4-846. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

