Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP
- PMID: 27530319
- PMCID: PMC4987667
- DOI: 10.1038/srep31729
Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP
Abstract
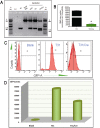
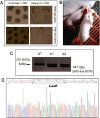
Predictable, clean genetic modification (GM) in livestock is important for reliable phenotyping and biosafety. Here we reported the generation of isozygous, functional myostatin (MSTN) knockout cloned pigs free of selectable marker gene (SMG) by CRISPR/Cas9 and Cre/LoxP. CRISPR/Cas9-mediated homologous recombination (HR) was exploited to knock out (KO) one allele of MSTN in pig primary cells. Cre recombinase was then used to excise the SMG with an efficiency of 82.7%. The SMG-free non-EGFP cells were isolated by flow cytometery and immediately used as donor nuclei for nuclear transfer. A total of 685 reconstructed embryos were transferred into three surrogates with one delivering two male live piglets. Molecular testing verified the mono-allelic MSTN KO and SMG deletion in these cloned pigs. Western blots showed approximately 50% decrease in MSTN and concurrent increased expression of myogenic genes in muscle. Histological examination revealed the enhanced myofiber quantity but myofiber size remained unaltered. Ultrasonic detection showed the increased longissimus muscle size and decreased backfat thickness. Precision editing of pig MSTN gene has generated isozygous, SMG-free MSTN KO cloned founders, which guaranteed a reliable route for elite livestock production and a strategy to minimize potential biological risks.
Figures


References
-
- Carlson D. F. et al. Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Research 20, 1125–1137 (2011). - PubMed
-
- Han H. et al. One-step generation of myostatin gene knockout sheep via the CRISPR/Cas9 system. Frontiers of Agricultural Science and Engineering 1, 2–5 (2014).
-
- Xu Y. et al. Excision of selectable genes from transgenic goat cells by a protein transducible TAT-Cre recombinase. Gene 419, 70–74 (2008). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

