A Chemical Probe for the ATAD2 Bromodomain
- PMID: 27530368
- PMCID: PMC7314595
- DOI: 10.1002/anie.201603928
A Chemical Probe for the ATAD2 Bromodomain
Abstract
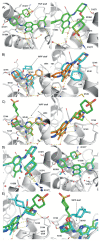
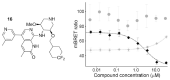
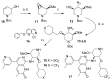
ATAD2 is a cancer-associated protein whose bromodomain has been described as among the least druggable of that target class. Starting from a potent lead, permeability and selectivity were improved through a dual approach: 1) using CF2 as a sulfone bio-isostere to exploit the unique properties of fluorine, and 2) using 1,3-interactions to control the conformation of a piperidine ring. This resulted in the first reported low-nanomolar, selective and cell permeable chemical probe for ATAD2.
Keywords: bioisosteres; conformation analysis; epigenetics; fluorine; medicinal chemistry.
© 2016 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures
Similar articles
-
Aiming to Miss a Moving Target: Bromo and Extra Terminal Domain (BET) Selectivity in Constrained ATAD2 Inhibitors.J Med Chem. 2018 Sep 27;61(18):8321-8336. doi: 10.1021/acs.jmedchem.8b00862. Epub 2018 Sep 18. J Med Chem. 2018. PMID: 30226378
-
Identification of a novel ligand for the ATAD2 bromodomain with selectivity over BRD4 through a fragment growing approach.Org Biomol Chem. 2018 Mar 14;16(11):1843-1850. doi: 10.1039/c8ob00099a. Org Biomol Chem. 2018. PMID: 29469144 Free PMC article.
-
Coordination of Di-Acetylated Histone Ligands by the ATAD2 Bromodomain.Int J Mol Sci. 2021 Aug 24;22(17):9128. doi: 10.3390/ijms22179128. Int J Mol Sci. 2021. PMID: 34502039 Free PMC article.
-
ATAD2 in cancer: a pharmacologically challenging but tractable target.Expert Opin Ther Targets. 2018 Jan;22(1):85-96. doi: 10.1080/14728222.2018.1406921. Epub 2017 Nov 23. Expert Opin Ther Targets. 2018. PMID: 29148850 Review.
-
Malignant genome reprogramming by ATAD2.Biochim Biophys Acta. 2013 Oct;1829(10):1010-4. doi: 10.1016/j.bbagrm.2013.06.003. Epub 2013 Jul 3. Biochim Biophys Acta. 2013. PMID: 23831842 Review.
Cited by
-
Androgen Receptor Deregulation Drives Bromodomain-Mediated Chromatin Alterations in Prostate Cancer.Cell Rep. 2017 Jun 6;19(10):2045-2059. doi: 10.1016/j.celrep.2017.05.049. Cell Rep. 2017. PMID: 28591577 Free PMC article.
-
Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.J Med Chem. 2022 Nov 24;65(22):15416-15432. doi: 10.1021/acs.jmedchem.2c01357. Epub 2022 Nov 11. J Med Chem. 2022. PMID: 36367089 Free PMC article.
-
GSK973 Is an Inhibitor of the Second Bromodomains (BD2s) of the Bromodomain and Extra-Terminal (BET) Family.ACS Med Chem Lett. 2020 Jul 6;11(8):1581-1587. doi: 10.1021/acsmedchemlett.0c00247. eCollection 2020 Aug 13. ACS Med Chem Lett. 2020. PMID: 32832027 Free PMC article.
-
Bromodomain inhibitors and therapeutic applications.Curr Opin Chem Biol. 2023 Aug;75:102323. doi: 10.1016/j.cbpa.2023.102323. Epub 2023 May 17. Curr Opin Chem Biol. 2023. PMID: 37207401 Free PMC article. Review.
-
Functional Roles of Bromodomain Proteins in Cancer.Cancers (Basel). 2021 Jul 19;13(14):3606. doi: 10.3390/cancers13143606. Cancers (Basel). 2021. PMID: 34298819 Free PMC article. Review.
References
-
- Caron C, et al. Oncogene. 2010;29:5171. - PubMed
-
- Boussouar F, Jamshidikia M, Morozumi Y, Rousseaux S, Khochbin S. Biochim Biophys Acta Gene Regul Mech. 2013;1829:1010. - PubMed
-
- Zou JX, Guo L, Revenko AS, Tepper CG, Gemo AT, Kung HJ, Chen HW. Cancer Res. 2009;69:3339. - PubMed
-
- Demont EH, et al. J Med Chem. 2015;58:5649. - PubMed
-
- Bamborough P, et al. J Med Chem. 2015;58:6151. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

