Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing
- PMID: 27530427
- PMCID: PMC5041492
- DOI: 10.1093/nar/gkw714
Structure of a human pre-40S particle points to a role for RACK1 in the final steps of 18S rRNA processing
Abstract
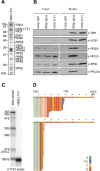
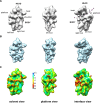
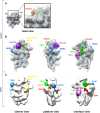
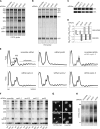
Synthesis of ribosomal subunits in eukaryotes is a complex and tightly regulated process that has been mostly characterized in yeast. The discovery of a growing number of diseases linked to defects in ribosome biogenesis calls for a deeper understanding of these mechanisms and of the specificities of human ribosome maturation. We present the 19 Å resolution cryo-EM reconstruction of a cytoplasmic precursor to the human small ribosomal subunit, purified by using the tagged ribosome biogenesis factor LTV1 as bait. Compared to yeast pre-40S particles, this first three-dimensional structure of a human 40S subunit precursor shows noticeable differences with respect to the position of ribosome biogenesis factors and uncovers the early deposition of the ribosomal protein RACK1 during subunit maturation. Consistently, RACK1 is required for efficient processing of the 18S rRNA 3'-end, which might be related to its role in translation initiation. This first structural analysis of a human pre-ribosomal particle sets the grounds for high-resolution studies of conformational transitions accompanying ribosomal subunit maturation.
© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Structural basis for the final steps of human 40S ribosome maturation.Nature. 2020 Nov;587(7835):683-687. doi: 10.1038/s41586-020-2929-x. Epub 2020 Nov 18. Nature. 2020. PMID: 33208940
-
Affinity grid-based cryo-EM of PKC binding to RACK1 on the ribosome.J Struct Biol. 2013 Feb;181(2):190-4. doi: 10.1016/j.jsb.2012.11.006. Epub 2012 Dec 8. J Struct Biol. 2013. PMID: 23228487 Free PMC article.
-
Conformational switches control early maturation of the eukaryotic small ribosomal subunit.Elife. 2019 Jun 17;8:e45185. doi: 10.7554/eLife.45185. Elife. 2019. PMID: 31206356 Free PMC article.
-
Maturation of pre-40S particles in yeast and humans.Wiley Interdiscip Rev RNA. 2019 Jan;10(1):e1516. doi: 10.1002/wrna.1516. Epub 2018 Nov 8. Wiley Interdiscip Rev RNA. 2019. PMID: 30406965 Review.
-
Assembly of the small ribosomal subunit in yeast: mechanism and regulation.RNA. 2018 Jul;24(7):881-891. doi: 10.1261/rna.066985.118. Epub 2018 Apr 30. RNA. 2018. PMID: 29712726 Free PMC article. Review.
Cited by
-
Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing.Nucleic Acids Res. 2019 Feb 28;47(4):1880-1895. doi: 10.1093/nar/gky1261. Nucleic Acids Res. 2019. PMID: 30576513 Free PMC article.
-
Neuronal ribosomes exhibit dynamic and context-dependent exchange of ribosomal proteins.Nat Commun. 2021 Oct 21;12(1):6127. doi: 10.1038/s41467-021-26365-x. Nat Commun. 2021. PMID: 34675203 Free PMC article.
-
Uncovering the assembly pathway of human ribosomes and its emerging links to disease.EMBO J. 2019 Jul 1;38(13):e100278. doi: 10.15252/embj.2018100278. Epub 2019 May 14. EMBO J. 2019. PMID: 31268599 Free PMC article. Review.
-
USP16 counteracts mono-ubiquitination of RPS27a and promotes maturation of the 40S ribosomal subunit.Elife. 2020 Mar 4;9:e54435. doi: 10.7554/eLife.54435. Elife. 2020. PMID: 32129764 Free PMC article.
-
RACK1 on and off the ribosome.RNA. 2019 Jul;25(7):881-895. doi: 10.1261/rna.071217.119. Epub 2019 Apr 25. RNA. 2019. PMID: 31023766 Free PMC article.
References
-
- Anger A.M., Armache J.-P., Berninghausen O., Habeck M., Subklewe M., Wilson D.N., Beckmann R. Structures of the human and Drosophila 80S ribosome. Nature. 2013;497:80–85. - PubMed
-
- Zemp I., Kutay U. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 2007;581:2783–2793. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

