The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3
- PMID: 27530752
- PMCID: PMC5890657
- DOI: 10.1093/jac/dkw321
The spread of KPC-producing Enterobacteriaceae in Spain: WGS analysis of the emerging high-risk clones of Klebsiella pneumoniae ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3
Abstract
Objectives: We analysed the microbiological traits and population structure of KPC-producing Enterobacteriaceae isolates collected in Spain between 2012 and 2014. We also performed a comparative WGS analysis of the three major KPC-producing Klebsiella pneumoniae clones detected.
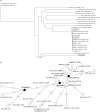
Methods: Carbapenemase and ESBL genes were sequenced. The Institut Pasteur MLST scheme was used. WGS data were used to construct phylogenetic trees, to identify the determinants of resistance and to de novo assemble the genome of one representative isolate of each of the three major K. pneumoniae clones.
Results: Of the 2443 carbapenemase-producing Enterobacteriaceae isolates identified during the study period, 111 (4.5%) produced KPC. Of these, 81 (73.0%) were K. pneumoniae and 13 (11.7%) were Enterobacter cloacae. Three major epidemic clones of K. pneumoniae were identified: ST11/KPC-2, ST101/KPC-2 and ST512/KPC-3. ST11/KPC-2 differed from ST101/KPC-2 and ST512/KPC-3 by 27 819 and 6924 SNPs, respectively. ST101/KPC-2 differed from ST512/KPC-3 by 28 345 SNPs. Nine acquired resistance genes were found in ST11/KPC-2, 11 in ST512/KPC-3 and 13 in ST101/KPC-2. ST101/KPC-2 had the highest number of virulence genes (20). An 11 bp deletion at the end of the mgrB sequence was the cause of colistin resistance in ST512/KPC-3.
Conclusions: KPC-producing Enterobacteriaceae are increasing in Spain. Most KPC-producing K. pneumoniae isolates belonged to only five clones: ST11 and ST512 caused interregional spread, ST101 caused regional spread and ST1961 and ST678 produced independent hospital outbreaks. ST101/KPC-2 had the highest number of resistance and virulence genes. ST101/KPC-2 and ST512/KPC-3 were recently implicated in the spread of KPC in Italy.
© The Author 2016. Published by Oxford University Press on behalf of the British Society for Antimicrobial Chemotherapy. All rights reserved. For Permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriaceae worldwide. Clin Microbiol Infect 2014; 20: 821–30. - PubMed
-
- Cantón R, Akóva M, Carmeli Y et al. . Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect 2012; 18: 413–31. - PubMed
-
- Oteo J, Miró E, Pérez-Vázquez M et al. . Evolution of carbapenemase-producing Enterobacteriaceae at the global and national level: what should be expected in the future? Enferm Infecc Microbiol Clin 2014; 32Suppl 4: 17–23. - PubMed
-
- Glasner C, Albiger B, Buist G et al. . Carbapenemase-producing Enterobacteriaceae in Europe: a survey among national experts from 39 countries, February 2013. Euro Surveill 2013; 18: pii=20525. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

