Telomeric RNAs are essential to maintain telomeres
- PMID: 27531349
- PMCID: PMC4992061
- DOI: 10.1038/ncomms12534
Telomeric RNAs are essential to maintain telomeres
Abstract
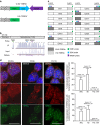
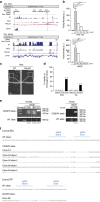
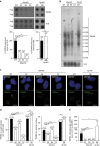
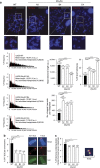
Telomeres are transcribed generating long non-coding RNAs known as TERRA. Deciphering the role of TERRA has been one of the unsolved issues of telomere biology in the past decade. This has been, in part, due to lack of knowledge on the TERRA loci, thus preventing functional genetic studies. Here, we describe that long non-coding RNAs with TERRA features are transcribed from the human 20q and Xp subtelomeres. Deletion of the 20q locus by using the CRISPR-Cas9 technology causes a dramatic decrease in TERRA levels, while deletion of the Xp locus does not result in decreased TERRA levels. Strikingly, 20q-TERRA ablation leads to dramatic loss of telomere sequences and the induction of a massive DNA damage response. These findings identify chromosome 20q as a main TERRA locus in human cells and represent the first demonstration in any organism of the essential role of TERRA in the maintenance of telomeres.
Figures

Comment in
-
Repurposing drugs to treat neurological diseases.J Neurol. 2018 Feb;265(2):446-448. doi: 10.1007/s00415-018-8732-z. J Neurol. 2018. PMID: 29322257 Free PMC article. No abstract available.
References
-
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E. & Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 (2007). - PubMed
-
- Schoeftner S. & Blasco M. A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 (2008). - PubMed
-
- Lopez de Silanes I., Stagno d'Alcontres M. & Blasco M. A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 1, 33 (2010). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

