Cytoplasmic dynein binding, run length, and velocity are guided by long-range electrostatic interactions
- PMID: 27531742
- PMCID: PMC4987762
- DOI: 10.1038/srep31523
Cytoplasmic dynein binding, run length, and velocity are guided by long-range electrostatic interactions
Abstract
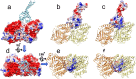
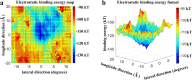
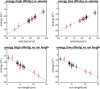
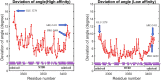
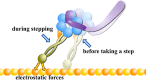
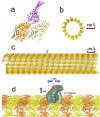
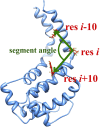
Dyneins are important molecular motors involved in many essential biological processes, including cargo transport along microtubules, mitosis, and in cilia. Dynein motility involves the coupling of microtubule binding and unbinding to a change in the configuration of the linker domain induced by ATP hydrolysis, which occur some 25 nm apart. This leaves the accuracy of dynein stepping relatively inaccurate and susceptible to thermal noise. Using multi-scale modeling with a computational focusing technique, we demonstrate that the microtubule forms an electrostatic funnel that guides the dynein's microtubule binding domain (MTBD) as it finally docks to the precise, keyed binding location on the microtubule. Furthermore, we demonstrate that electrostatic component of the MTBD's binding free energy is linearly correlated with the velocity and run length of dynein, and we use this linearity to predict the effect of mutating each glutamic and aspartic acid located in MTBD domain to alanine. Lastly, we show that the binding of dynein to the microtubule is associated with conformational changes involving several helices, and we localize flexible hinge points within the stalk helices. Taken all together, we demonstrate that long range electrostatic interactions bring a level of precision to an otherwise noisy dynein stepping process.
Figures

References
-
- King S. M. & Stephen K. M. Dyneins: Structure, Biology and Disease. (Academic Press, 2011).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

