'Cytology-on-a-chip' based sensors for monitoring of potentially malignant oral lesions
- PMID: 27531880
- PMCID: PMC5056560
- DOI: 10.1016/j.oraloncology.2016.07.002
'Cytology-on-a-chip' based sensors for monitoring of potentially malignant oral lesions
Abstract
Despite significant advances in surgical procedures and treatment, long-term prognosis for patients with oral cancer remains poor, with survival rates among the lowest of major cancers. Better methods are desperately needed to identify potential malignancies early when treatments are more effective.
Objective: To develop robust classification models from cytology-on-a-chip measurements that mirror diagnostic performance of gold standard approach involving tissue biopsy.
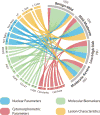
Materials and methods: Measurements were recorded from 714 prospectively recruited patients with suspicious lesions across 6 diagnostic categories (each confirmed by tissue biopsy -histopathology) using a powerful new 'cytology-on-a-chip' approach capable of executing high content analysis at a single cell level. Over 200 cellular features related to biomarker expression, nuclear parameters and cellular morphology were recorded per cell. By cataloging an average of 2000 cells per patient, these efforts resulted in nearly 13 million indexed objects.
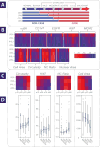
Results: Binary "low-risk"/"high-risk" models yielded AUC values of 0.88 and 0.84 for training and validation models, respectively, with an accompanying difference in sensitivity+specificity of 6.2%. In terms of accuracy, this model accurately predicted the correct diagnosis approximately 70% of the time, compared to the 69% initial agreement rate of the pool of expert pathologists. Key parameters identified in these models included cell circularity, Ki67 and EGFR expression, nuclear-cytoplasmic ratio, nuclear area, and cell area.
Conclusions: This chip-based approach yields objective data that can be leveraged for diagnosis and management of patients with PMOL as well as uncovering new molecular-level insights behind cytological differences across the OED spectrum.
Keywords: Cytology; High content analysis; LASSO; Machine learning; Microfluidic; Oral cancer; Oral epithelial dysplasia; Random forest.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Principal Investigator, John T. McDevitt, has an equity interest in SensoDX, LLC. and also serves on their Scientific Advisory Board. The terms of this arrangement have been reviewed and approved by New York University in accordance with its conflict of interest policies.
Figures


References
-
- Shulman JD, Beach MM, Rivera-Hidalgo F. The prevalence of oral mucosal lesions in U.S. adults: Data from the Third National Health and Nutrition Examination Survey, 1988–1994. J Am Dent Assoc. 2004;135:1279–86. - PubMed
-
- Lee JJ, Hong WK, Hittelman WN, Mao L, Lotan R, Shin DM, et al. Predicting cancer development in oral leukoplakia: Ten years of translational research. Clin Cancer Res. 2000;6:1702–10. - PubMed
-
- Lydiatt DD. Cancer of the oral cavity and medical malpractice. Laryngoscope. 2002;112:816–9. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

