The Stringent Response Promotes Antibiotic Resistance Dissemination by Regulating Integron Integrase Expression in Biofilms
- PMID: 27531906
- PMCID: PMC4992968
- DOI: 10.1128/mBio.00868-16
The Stringent Response Promotes Antibiotic Resistance Dissemination by Regulating Integron Integrase Expression in Biofilms
Abstract
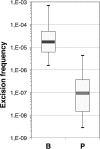
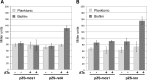
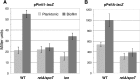
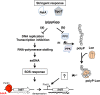
Class 1 integrons are genetic systems that enable bacteria to capture and express gene cassettes. These integrons, when isolated in clinical contexts, most often carry antibiotic resistance gene cassettes. They play a major role in the dissemination of antibiotic resistance among Gram-negative bacteria. The key element of integrons is the integrase, which allows gene cassettes to be acquired and shuffled. Planktonic culture experiments have shown that integrase expression is regulated by the bacterial SOS response. In natural settings, however, bacteria generally live in biofilms, which are characterized by strong antibiotic resilience and by increased expression of stress-related genes. Here, we report that under biofilm conditions, the stringent response, which is induced upon starvation, (i) increases basal integrase and SOS regulon gene expression via induction of the SOS response and (ii) exerts biofilm-specific regulation of the integrase via the Lon protease. This indicates that biofilm environments favor integron-mediated acquisition of antibiotic resistance and other adaptive functions encoded by gene cassettes.
Importance: Multidrug-resistant bacteria are becoming a worldwide health problem. Integrons are bacterial genetic platforms that allow the bacteria to capture and express gene cassettes. In clinical settings, integrons play a major role in the dissemination of antibiotic resistance gene cassettes among Gram-negative bacteria. Cassette capture is catalyzed by the integron integrase, whose expression is induced by DNA damage and controlled by the bacterial SOS response in laboratory planktonic cultures. In natural settings, bacteria usually grow in heterogeneous environments known as biofilms, which have very different conditions than planktonic cultures. Integrase regulation has not been investigated in biofilms. Our results showed that in addition to the SOS response, the stringent response (induced upon starvation) is specifically involved in the regulation of class 1 integron integrases in biofilms. This study shows that biofilms are favorable environments for integron-mediated acquisition/exchange of antibiotic resistance genes by bacteria and for the emergence of multidrug-resistant bacteria.
Copyright © 2016 Strugeon et al.
Figures
Similar articles
-
Class 1 integrons are low-cost structures in Escherichia coli.ISME J. 2017 Jul;11(7):1535-1544. doi: 10.1038/ismej.2017.38. Epub 2017 Apr 7. ISME J. 2017. PMID: 28387772 Free PMC article.
-
Diversity of Class 1 Integron Gene Cassette Rearrangements Selected under Antibiotic Pressure.J Bacteriol. 2015 Jul;197(13):2171-2178. doi: 10.1128/JB.02455-14. Epub 2015 Apr 20. J Bacteriol. 2015. PMID: 25897031 Free PMC article.
-
Activation of class 1 integron integrase is promoted in the intestinal environment.PLoS Genet. 2022 Apr 28;18(4):e1010177. doi: 10.1371/journal.pgen.1010177. eCollection 2022 Apr. PLoS Genet. 2022. PMID: 35482826 Free PMC article.
-
Epidemiology and molecular mechanism of integron-mediated antibiotic resistance in Shigella.Arch Microbiol. 2011 Nov;193(11):767-74. doi: 10.1007/s00203-011-0744-3. Epub 2011 Aug 14. Arch Microbiol. 2011. PMID: 21842348 Review.
-
Integron and its role in antimicrobial resistance: A literature review on some bacterial pathogens.Iran J Basic Med Sci. 2021 Feb;24(2):136-142. doi: 10.22038/ijbms.2020.48905.11208. Iran J Basic Med Sci. 2021. PMID: 33953851 Free PMC article. Review.
Cited by
-
The fate of sulfonamide resistance genes and anthropogenic pollution marker intI1 after discharge of wastewater into a pristine river stream.Front Microbiol. 2023 Jan 25;14:1058350. doi: 10.3389/fmicb.2023.1058350. eCollection 2023. Front Microbiol. 2023. PMID: 36760511 Free PMC article.
-
Unbridled Integrons: A Matter of Host Factors.Cells. 2022 Mar 8;11(6):925. doi: 10.3390/cells11060925. Cells. 2022. PMID: 35326376 Free PMC article. Review.
-
Microbiota of Chronic Diabetic Wounds: Ecology, Impact, and Potential for Innovative Treatment Strategies.Front Microbiol. 2017 Sep 21;8:1791. doi: 10.3389/fmicb.2017.01791. eCollection 2017. Front Microbiol. 2017. PMID: 28983285 Free PMC article. Review.
-
Inorganic polyphosphate and the stringent response coordinately control cell division and cell morphology in Escherichia coli.mBio. 2025 Feb 5;16(2):e0351124. doi: 10.1128/mbio.03511-24. Epub 2024 Dec 27. mBio. 2025. PMID: 39727417 Free PMC article.
-
Phenotypic and Multi-Omics Characterization of Escherichia coli K-12 Adapted to Chlorhexidine Identifies the Role of MlaA and Other Cell Envelope Alterations Regulated by Stress Inducible Pathways in CHX Resistance.Front Mol Biosci. 2021 May 19;8:659058. doi: 10.3389/fmolb.2021.659058. eCollection 2021. Front Mol Biosci. 2021. PMID: 34095221 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

