Comparison of Diagnostic Algorithms for Detecting Toxigenic Clostridium difficile in Routine Practice at a Tertiary Referral Hospital in Korea
- PMID: 27532104
- PMCID: PMC4988646
- DOI: 10.1371/journal.pone.0161139
Comparison of Diagnostic Algorithms for Detecting Toxigenic Clostridium difficile in Routine Practice at a Tertiary Referral Hospital in Korea
Abstract
Since every single test has some limitations for detecting toxigenic Clostridium difficile, multistep algorithms are recommended. This study aimed to compare the current, representative diagnostic algorithms for detecting toxigenic C. difficile, using VIDAS C. difficile toxin A&B (toxin ELFA), VIDAS C. difficile GDH (GDH ELFA, bioMérieux, Marcy-l'Etoile, France), and Xpert C. difficile (Cepheid, Sunnyvale, California, USA). In 271 consecutive stool samples, toxigenic culture, toxin ELFA, GDH ELFA, and Xpert C. difficile were performed. We simulated two algorithms: screening by GDH ELFA and confirmation by Xpert C. difficile (GDH + Xpert) and combined algorithm of GDH ELFA, toxin ELFA, and Xpert C. difficile (GDH + Toxin + Xpert). The performance of each assay and algorithm was assessed. The agreement of Xpert C. difficile and two algorithms (GDH + Xpert and GDH+ Toxin + Xpert) with toxigenic culture were strong (Kappa, 0.848, 0.857, and 0.868, respectively). The sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of algorithms (GDH + Xpert and GDH + Toxin + Xpert) were 96.7%, 95.8%, 85.0%, 98.1%, and 94.5%, 95.8%, 82.3%, 98.5%, respectively. There were no significant differences between Xpert C. difficile and two algorithms in sensitivity, specificity, PPV and NPV. The performances of both algorithms for detecting toxigenic C. difficile were comparable to that of Xpert C. difficile. Either algorithm would be useful in clinical laboratories and can be optimized in the diagnostic workflow of C. difficile depending on costs, test volume, and clinical needs.
Conflict of interest statement
Figures
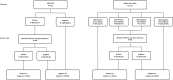
References
-
- Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, et al. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the society for healthcare epidemiology of America (SHEA) and the infectious diseases society of America (IDSA). Infect Control Hosp Epidemiol. 2010;31(5):431–55. Epub 2010/03/24. 10.1086/651706 - DOI - PubMed
-
- Planche TD, Davies KA, Coen PG, Finney JM, Monahan IM, Morris KA, et al. Differences in outcome according to Clostridium difficile testing method: a prospective multicentre diagnostic validation study of C difficile infection. Lancet Infect Dis. 2013;13(11):936–45. Epub 2013/09/07. 10.1016/S1473-3099(13)70200-7 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

