Positive Selection Pressure Drives Variation on the Surface-Exposed Variable Proteins of the Pathogenic Neisseria
- PMID: 27532335
- PMCID: PMC5020929
- DOI: 10.1371/journal.pone.0161348
Positive Selection Pressure Drives Variation on the Surface-Exposed Variable Proteins of the Pathogenic Neisseria
Abstract
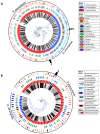
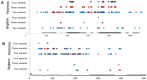
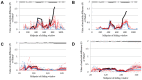
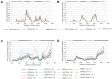
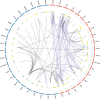
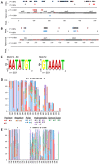
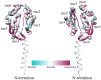
Pathogenic species of Neisseria utilize variable outer membrane proteins to facilitate infection and proliferation within the human host. However, the mechanisms behind the evolution of these variable alleles remain largely unknown due to analysis of previously limited datasets. In this study, we have expanded upon the previous analyses to substantially increase the number of analyzed sequences by including multiple diverse strains, from various geographic locations, to determine whether positive selective pressure is exerted on the evolution of these variable genes. Although Neisseria are naturally competent, this analysis indicates that only intrastrain horizontal gene transfer among the pathogenic Neisseria principally account for these genes exhibiting linkage equilibrium which drives the polymorphisms evidenced within these alleles. As the majority of polymorphisms occur across species, the divergence of these variable genes is dependent upon the species and is independent of geographical location, disease severity, or serogroup. Tests of neutrality were able to detect strong selection pressures acting upon both the opa and pil gene families, and were able to locate the majority of these sites within the exposed variable regions of the encoded proteins. Evidence of positive selection acting upon the hypervariable domains of Opa contradicts previous beliefs and provides evidence for selection of receptor binding. As the pathogenic Neisseria reside exclusively within the human host, the strong selection pressures acting upon both the opa and pil gene families provide support for host immune system pressure driving sequence polymorphisms within these variable genes.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

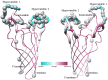
Similar articles
-
Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis.Mol Microbiol. 1994 Apr;12(2):171-80. doi: 10.1111/j.1365-2958.1994.tb01006.x. Mol Microbiol. 1994. PMID: 7520117
-
Frequency and rate of pilin antigenic variation of Neisseria meningitidis.J Bacteriol. 2010 Jul;192(14):3822-3. doi: 10.1128/JB.00280-10. Epub 2010 May 14. J Bacteriol. 2010. PMID: 20472803 Free PMC article.
-
Identification and characterization of specific sequences encoding pathogenicity associated proteins in the genome of commensal Neisseria species.FEMS Microbiol Lett. 1995 Jan 15;125(2-3):255-63. doi: 10.1111/j.1574-6968.1995.tb07366.x. FEMS Microbiol Lett. 1995. PMID: 7875573
-
Focusing homologous recombination: pilin antigenic variation in the pathogenic Neisseria.Mol Microbiol. 2011 Sep;81(5):1136-43. doi: 10.1111/j.1365-2958.2011.07773.x. Epub 2011 Aug 4. Mol Microbiol. 2011. PMID: 21812841 Free PMC article. Review.
-
Phase and antigenic variation of pili and outer membrane protein II of Neisseria gonorrhoeae.J Infect Dis. 1986 Feb;153(2):196-201. doi: 10.1093/infdis/153.2.196. J Infect Dis. 1986. PMID: 2418125 Review. No abstract available.
Cited by
-
Neisseria gonorrhoeae infects the heterogeneous epithelia of the human cervix using distinct mechanisms.PLoS Pathog. 2019 Dec 2;15(12):e1008136. doi: 10.1371/journal.ppat.1008136. eCollection 2019 Dec. PLoS Pathog. 2019. PMID: 31790511 Free PMC article.
-
The effect of recombination on the evolution of a population of Neisseria meningitidis.Genome Res. 2021 Jul;31(7):1258-1268. doi: 10.1101/gr.264465.120. Epub 2021 Jun 9. Genome Res. 2021. PMID: 34108268 Free PMC article.
-
Neisseria gonorrhoeae Aggregation Reduces Its Ceftriaxone Susceptibility.Antibiotics (Basel). 2018 Jun 15;7(2):48. doi: 10.3390/antibiotics7020048. Antibiotics (Basel). 2018. PMID: 29914058 Free PMC article.
-
In silico functional and evolutionary analyses of rubber oxygenases (RoxA and RoxB).3 Biotech. 2020 Sep;10(9):376. doi: 10.1007/s13205-020-02371-6. Epub 2020 Aug 5. 3 Biotech. 2020. PMID: 32802718 Free PMC article.
-
A homopolymeric adenosine tract in the promoter region of nspA influences factor H-mediated serum resistance in Neisseria meningitidis.Sci Rep. 2019 Feb 25;9(1):2736. doi: 10.1038/s41598-019-39231-0. Sci Rep. 2019. PMID: 30804422 Free PMC article.
References
-
- Scherp HW. Neisseria and Neisserial Infections. Annu Rev Microbiol. 1955;9: 319–334. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

