Iron Content Affects Lipogenic Gene Expression in the Muscle of Nelore Beef Cattle
- PMID: 27532424
- PMCID: PMC4988672
- DOI: 10.1371/journal.pone.0161160
Iron Content Affects Lipogenic Gene Expression in the Muscle of Nelore Beef Cattle
Abstract
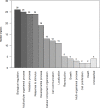
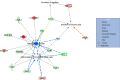
Iron (Fe) is an essential mineral for metabolism and plays a central role in a range of biochemical processes. Therefore, this study aimed to identify differentially expressed (DE) genes and metabolic pathways in Longissimus dorsi (LD) muscle from cattle with divergent iron content, as well as to investigate the likely role of these DE genes in biological processes underlying beef quality parameters. Samples for RNA extraction for sequencing and iron, copper, manganese, and zinc determination were collected from LD muscles at slaughter. Eight Nelore steers, with extreme genomic estimated breeding values for iron content (Fe-GEBV), were selected from a reference population of 373 animals. From the 49 annotated DE genes (FDR<0.05) found between the two groups, 18 were up-regulated and 31 down-regulated for the animals in the low Fe-GEBV group. The functional enrichment analyses identified several biological processes, such as lipid transport and metabolism, and cell growth. Lipid metabolism was the main pathway observed in the analysis of metabolic and canonical signaling pathways for the genes identified as DE, including the genes FASN, FABP4, and THRSP, which are functional candidates for beef quality, suggesting reduced lipogenic activities with lower iron content. Our results indicate metabolic pathways that are partially influenced by iron, contributing to a better understanding of its participation in skeletal muscle physiology.
Conflict of interest statement
Figures

References
-
- Beard JL. Iron biology in immune function, muscle metabolism and neuronal functioning. J Nutr. 2001;131: 568S–579S - PubMed
-
- Dauncey MJ, Katsumata M, White P. Nutrition, hormone receptor expression and gene interactions: implications for development and disease In: Pas MFW, Evertes ME, Haagsman HP, editors. Muscle development of livestock animals: physiology, genetics and meat quality. Wallingford: CABI; 2004. p. 419
-
- Choi YM, Kim BC. Muscle fiber characteristics, myofibrillar protein isoforms, and meat quality. Livest Sci. Elsevier B.V.; 2009;122: 105–118. 10.1016/j.livsci.2008.08.015 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

