Genetic Misdiagnoses and the Potential for Health Disparities
- PMID: 27532831
- PMCID: PMC5292722
- DOI: 10.1056/NEJMsa1507092
Genetic Misdiagnoses and the Potential for Health Disparities
Abstract
Background: For more than a decade, risk stratification for hypertrophic cardiomyopathy has been enhanced by targeted genetic testing. Using sequencing results, clinicians routinely assess the risk of hypertrophic cardiomyopathy in a patient's relatives and diagnose the condition in patients who have ambiguous clinical presentations. However, the benefits of genetic testing come with the risk that variants may be misclassified.
Methods: Using publicly accessible exome data, we identified variants that have previously been considered causal in hypertrophic cardiomyopathy and that are overrepresented in the general population. We studied these variants in diverse populations and reevaluated their initial ascertainments in the medical literature. We reviewed patient records at a leading genetic-testing laboratory for occurrences of these variants during the near-decade-long history of the laboratory.
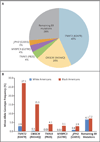
Results: Multiple patients, all of whom were of African or unspecified ancestry, received positive reports, with variants misclassified as pathogenic on the basis of the understanding at the time of testing. Subsequently, all reported variants were recategorized as benign. The mutations that were most common in the general population were significantly more common among black Americans than among white Americans (P<0.001). Simulations showed that the inclusion of even small numbers of black Americans in control cohorts probably would have prevented these misclassifications. We identified methodologic shortcomings that contributed to these errors in the medical literature.
Conclusions: The misclassification of benign variants as pathogenic that we found in our study shows the need for sequencing the genomes of diverse populations, both in asymptomatic controls and the tested patient population. These results expand on current guidelines, which recommend the use of ancestry-matched controls to interpret variants. As additional populations of different ancestry backgrounds are sequenced, we expect variant reclassifications to increase, particularly for ancestry groups that have historically been less well studied. (Funded by the National Institutes of Health.).
Figures
Comment in
-
Body Mass Index As a Measure of Obesity: Racial Differences in Predictive Value for Health Parameters During Pregnancy.J Womens Health (Larchmt). 2016 Dec;25(12):1198. doi: 10.1089/jwh.2016.6184. J Womens Health (Larchmt). 2016. PMID: 27982746 No abstract available.
References
-
- Maron BJ. Hypertrophic cardiomyopathy: a systematic review. JAMA. 2002;287:1308–1320. - PubMed
-
- Maron BJ, Gardin JM, Flack JM, Gidding SS, Kurosaki TT, Bild DE. Prevalence of hypertrophic cardiomyopathy in a general population of young adults: echocardiographic analysis of 4111 subjects in the CARDIA Study. Circulation. 1995;92:785–789. - PubMed
-
- Maron BJ, Maron MS. Hypertrophic cardiomyopathy. Lancet. 2013;381:242–255. - PubMed
-
- Maron BJ, Maron MS, Semsarian C. Genetics of hypertrophic cardiomyopathy after 20 years: clinical perspectives. J Am Coll Cardiol. 2012;60:705–715. - PubMed
-
- Weidemann F, Niemann M, Breunig F, et al. Long-term effects of enzyme replacement therapy on Fabry cardiomyopathy: evidence for a better outcome with early treatment. Circulation. 2009;119:524–529. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
