Computational Insights into Five- versus Six-Coordinate Iron Center in Ferrous Soybean Lipoxygenase
- PMID: 27532889
- PMCID: PMC5117133
- DOI: 10.1021/acs.jpclett.6b01626
Computational Insights into Five- versus Six-Coordinate Iron Center in Ferrous Soybean Lipoxygenase
Abstract
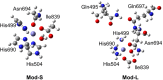
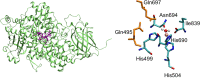
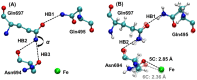
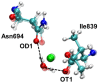
Soybean lipoxygenase (SLO) serves as a prototype for fundamental understanding of hydrogen tunneling in enzymes. Its reactivity depends on the active site structure around a mononuclear, nonheme iron center. The available crystal structures indicate five-coordinate iron, while magnetic circular dichroism experiments suggest significant populations of both five-coordinate (5C) and six-coordinate (6C) iron in ferrous SLO. Quantum mechanical calculations of gas phase models produce only 6C geometries. Herein mixed quantum mechanical/molecular mechanical (QM/MM) calculations are employed to identify and characterize the 5C and 6C geometries. These calculations highlight the importance of the protein environment, particularly two Gln residues in a hydrogen-bonding network with Asn694, the ligand that can dissociate. This hydrogen-bonding network is similar in both geometries, but twisting of a dihedral angle in Asn694 moves its oxygen away from the iron in the 5C geometry. These insights are important for future simulations of SLO.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Density-functional investigation on the mechanism of H-atom abstraction by lipoxygenase.J Biol Inorg Chem. 2003 Feb;8(3):294-305. doi: 10.1007/s00775-002-0415-6. Epub 2002 Nov 14. J Biol Inorg Chem. 2003. PMID: 12589565
-
Kinetic, spectroscopic, and structural investigations of the soybean lipoxygenase-1 first-coordination sphere mutant, Asn694Gly.Biochemistry. 2006 Aug 29;45(34):10233-42. doi: 10.1021/bi060577e. Biochemistry. 2006. PMID: 16922498
-
Spectroscopic characterization of soybean lipoxygenase-1 mutants: the role of second coordination sphere residues in the regulation of enzyme activity.Biochemistry. 2003 Jun 24;42(24):7294-302. doi: 10.1021/bi027380g. Biochemistry. 2003. PMID: 12809485
-
Understanding Biological Hydrogen Transfer Through the Lens of Temperature Dependent Kinetic Isotope Effects.Acc Chem Res. 2018 Sep 18;51(9):1966-1974. doi: 10.1021/acs.accounts.8b00226. Epub 2018 Aug 28. Acc Chem Res. 2018. PMID: 30152685 Free PMC article. Review.
-
Lipoxygenases: structural principles and spectroscopy.Annu Rev Biophys Biomol Struct. 1996;25:431-59. doi: 10.1146/annurev.bb.25.060196.002243. Annu Rev Biophys Biomol Struct. 1996. PMID: 8800477 Review.
Cited by
-
A Foundational Shift in Models for Enzyme Function.J Am Chem Soc. 2025 May 7;147(18):14884-14904. doi: 10.1021/jacs.5c02388. Epub 2025 Apr 25. J Am Chem Soc. 2025. PMID: 40277147 Free PMC article. Review.
-
13C ENDOR Spectroscopy of Lipoxygenase-Substrate Complexes Reveals the Structural Basis for C-H Activation by Tunneling.J Am Chem Soc. 2017 Feb 8;139(5):1984-1997. doi: 10.1021/jacs.6b11856. Epub 2017 Jan 25. J Am Chem Soc. 2017. PMID: 28121140 Free PMC article.
-
Characterization of the Fleeting Hydroxoiron(III) Complex of the Pentadentate TMC-py Ligand.Inorg Chem. 2017 Sep 18;56(18):11129-11140. doi: 10.1021/acs.inorgchem.7b01459. Epub 2017 Aug 31. Inorg Chem. 2017. PMID: 28858496 Free PMC article.
-
EPR Spectroscopic Studies of Lipoxygenases.Chem Asian J. 2020 Jan 2;15(1):42-50. doi: 10.1002/asia.201901461. Epub 2019 Dec 5. Chem Asian J. 2020. PMID: 31782616 Free PMC article. Review.
References
-
- Siedow J. N. Plant Lipoxygenase - Structure and Function. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 145–188. 10.1146/annurev.pp.42.060191.001045. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

