Digits and fin rays share common developmental histories
- PMID: 27533041
- PMCID: PMC5161576
- DOI: 10.1038/nature19322
Digits and fin rays share common developmental histories
Abstract
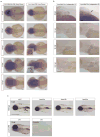
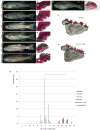
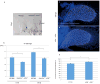
Understanding the evolutionary transformation of fish fins into tetrapod limbs is a fundamental problem in biology. The search for antecedents of tetrapod digits in fish has remained controversial because the distal skeletons of limbs and fins differ structurally, developmentally, and histologically. Moreover, comparisons of fins with limbs have been limited by a relative paucity of data on the cellular and molecular processes underlying the development of the fin skeleton. Here, we provide a functional analysis, using CRISPR/Cas9 and fate mapping, of 5' hox genes and enhancers in zebrafish that are indispensable for the development of the wrists and digits of tetrapods. We show that cells marked by the activity of an autopodial hoxa13 enhancer exclusively form elements of the fin fold, including the osteoblasts of the dermal rays. In hox13 knockout fish, we find that a marked reduction and loss of fin rays is associated with an increased number of endochondral distal radials. These discoveries reveal a cellular and genetic connection between the fin rays of fish and the digits of tetrapods and suggest that digits originated via the transition of distal cellular fates.
Figures






Comment in
-
Evolutionary biology: Fin to limb within our grasp.Nature. 2016 Sep 8;537(7619):176-177. doi: 10.1038/nature19425. Epub 2016 Aug 17. Nature. 2016. PMID: 27533039 No abstract available.
References
-
- Clack JA. Gaining Ground: The Origin and Evolution of Tetrapods. Indiana University Press; Indiana: 2012.
-
- Clack JA. The Fin to Limb Transition: New Data, Interpretations, and Hypotheses from Paleontology and Developmental Biology. 2009;37:163–179.
-
- Schneider I, Shubin NH. The origin of the tetrapod limb: from expeditions to enhancers. Trends Genet. 2013;29:419–26. - PubMed
-
- Fromental-Ramain C, et al. Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development. 1996;122:2997–3011. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

