The effect of the lamin A and its mutants on nuclear structure, cell proliferation, protein stability, and mobility in embryonic cells
- PMID: 27534416
- PMCID: PMC5509783
- DOI: 10.1007/s00412-016-0610-9
The effect of the lamin A and its mutants on nuclear structure, cell proliferation, protein stability, and mobility in embryonic cells
Abstract
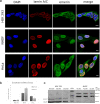
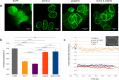
LMNA gene encodes for nuclear intermediate filament proteins lamin A/C. Mutations in this gene lead to a spectrum of genetic disorders, collectively referred to as laminopathies. Lamin A/C are widely expressed in most differentiated somatic cells but not in early embryos and some undifferentiated cells. To investigate the role of lamin A/C in cell phenotype maintenance and differentiation, which could be a determinant of the pathogenesis of laminopathies, we examined the role played by exogenous lamin A and its mutants in differentiated cell lines (HeLa, NHDF) and less-differentiated HEK 293 cells. We introduced exogenous wild-type and mutated (H222P, L263P, E358K D446V, and ∆50) lamin A into different cell types and analyzed proteins' impact on proliferation, protein mobility, and endogenous nuclear envelope protein distribution. The mutants give rise to a broad spectrum of nuclear phenotypes and relocate lamin C. The mutations ∆50 and D446V enhance proliferation in comparison to wild-type lamin A and control cells, but no changes in exogenous protein mobility measured by FRAP were observed. Interestingly, although transcripts for lamins A and C are at similar level in HEK 293 cells, only lamin C protein is detected in western blots. Also, exogenous lamin A and its mutants, when expressed in HEK 293 cells underwent posttranscriptional processing. Overall, our results provide new insight into the maintenance of lamin A in less-differentiated cells. Embryonic cells are very sensitive to lamin A imbalance, and its upregulation disturbs lamin C, which may influence gene expression and many regulatory pathways.
Keywords: Emery-Dreifuss muscular dystrophy; Human embryonic kidney 293; Hutchinson-Gilford progeria syndrome; Lamin A/C; Laminopathy; Nuclear envelope.
Conflict of interest statement
Funding
This study was supported by the Wroclaw Research Center EIT+ under the project Biotechnologies and Advanced Medical Technologies BioMed (POIG.01.01.02–02–003/08) from the Regional Developmental Fund. KP received funds from the National Science Centre (doctorate scholarship ETIUDA UMO-2014/12/T/NZ3/00504). The preparation and publication of the manuscript was supported with the KNOW (Country Leading Scientific Center for Biotechnology) grant from the Polish Ministry of Science.
Conflict of interest
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
Quantitative analysis of localization and nuclear aggregate formation induced by GFP-lamin A mutant proteins in living HeLa cells.J Cell Biochem. 2006 Jul 1;98(4):810-26. doi: 10.1002/jcb.20791. J Cell Biochem. 2006. PMID: 16440304
-
Specific contribution of lamin A and lamin C in the development of laminopathies.Exp Cell Res. 2008 Aug 1;314(13):2362-75. doi: 10.1016/j.yexcr.2008.04.017. Epub 2008 May 10. Exp Cell Res. 2008. PMID: 18538321 Free PMC article.
-
Lamins A and C are differentially dysfunctional in autosomal dominant Emery-Dreifuss muscular dystrophy.Eur J Cell Biol. 2005 Sep;84(9):765-81. doi: 10.1016/j.ejcb.2005.04.004. Eur J Cell Biol. 2005. PMID: 16218190
-
The laminopathies: a clinical review.Clin Genet. 2006 Oct;70(4):261-74. doi: 10.1111/j.1399-0004.2006.00677.x. Clin Genet. 2006. PMID: 16965317 Review.
-
Lamin A/C Mechanotransduction in Laminopathies.Cells. 2020 May 24;9(5):1306. doi: 10.3390/cells9051306. Cells. 2020. PMID: 32456328 Free PMC article. Review.
Cited by
-
The Nuclear Lamina: Protein Accumulation and Disease.Biomedicines. 2020 Jul 1;8(7):188. doi: 10.3390/biomedicines8070188. Biomedicines. 2020. PMID: 32630170 Free PMC article. Review.
-
Inhibition of FAK Signaling Elicits Lamin A/C-Associated Nuclear Deformity and Cellular Senescence.Front Oncol. 2019 Jan 30;9:22. doi: 10.3389/fonc.2019.00022. eCollection 2019. Front Oncol. 2019. PMID: 30761269 Free PMC article.
-
Drosophila Models Reveal Properties of Mutant Lamins That Give Rise to Distinct Diseases.Cells. 2023 Apr 12;12(8):1142. doi: 10.3390/cells12081142. Cells. 2023. PMID: 37190051 Free PMC article.
-
The E262K mutation in Lamin A links nuclear proteostasis imbalance to laminopathy-associated premature aging.Aging Cell. 2022 Nov;21(11):e13688. doi: 10.1111/acel.13688. Epub 2022 Oct 12. Aging Cell. 2022. PMID: 36225129 Free PMC article.
-
Emerin Is Required for Proper Nucleus Reassembly after Mitosis: Implications for New Pathogenetic Mechanisms for Laminopathies Detected in EDMD1 Patients.Cells. 2019 Mar 13;8(3):240. doi: 10.3390/cells8030240. Cells. 2019. PMID: 30871242 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

