Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia
- PMID: 27535106
- PMCID: PMC5584808
- DOI: 10.1158/2159-8290.CD-16-0237
Targeting Chromatin Regulators Inhibits Leukemogenic Gene Expression in NPM1 Mutant Leukemia
Abstract
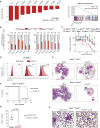
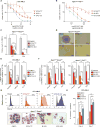
Homeobox (HOX) proteins and the receptor tyrosine kinase FLT3 are frequently highly expressed and mutated in acute myeloid leukemia (AML). Aberrant HOX expression is found in nearly all AMLs that harbor a mutation in the Nucleophosmin (NPM1) gene, and FLT3 is concomitantly mutated in approximately 60% of these cases. Little is known about how mutant NPM1 (NPM1mut) cells maintain aberrant gene expression. Here, we demonstrate that the histone modifiers MLL1 and DOT1L control HOX and FLT3 expression and differentiation in NPM1mut AML. Using a CRISPR/Cas9 genome editing domain screen, we show NPM1mut AML to be exceptionally dependent on the menin binding site in MLL1. Pharmacologic small-molecule inhibition of the menin-MLL1 protein interaction had profound antileukemic activity in human and murine models of NPM1mut AML. Combined pharmacologic inhibition of menin-MLL1 and DOT1L resulted in dramatic suppression of HOX and FLT3 expression, induction of differentiation, and superior activity against NPM1mut leukemia.
Significance: MLL1 and DOT1L are chromatin regulators that control HOX, MEIS1, and FLT3 expression and are therapeutic targets in NPM1mut AML. Combinatorial small-molecule inhibition has synergistic on-target activity and constitutes a novel therapeutic concept for this common AML subtype. Cancer Discov; 6(10); 1166-81. ©2016 AACR.See related commentary by Hourigan and Aplan, p. 1087This article is highlighted in the In This Issue feature, p. 1069.
©2016 American Association for Cancer Research.
Figures





Comment in
-
Accurate Medicine: Indirect Targeting of NPM1-Mutated AML.Cancer Discov. 2016 Oct;6(10):1087-1089. doi: 10.1158/2159-8290.CD-16-0917. Cancer Discov. 2016. PMID: 27698101 Free PMC article.
References
-
- Giampaolo A, Sterpetti P, Bulgarini D, Samoggia P, Pelosi E, Valtieri M, et al. Key functional role and lineage-specific expression of selected HOXB genes in purified hematopoietic progenitor differentiation. Blood. 1994 Dec 1;84(11):3637–47. - PubMed
-
- Alharbi RA, Pettengell R, Pandha HS, Morgan R. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 2013 Apr;27(5):1000–8. - PubMed
-
- Argiropoulos B, Humphries RK. Hox genes in hematopoiesis and leukemogenesis. Oncogene. 2007 Oct 15;26(47):6766–76. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

