Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method
- PMID: 27535322
- PMCID: PMC4989495
- DOI: 10.1038/srep32141
Direct visualization of both DNA and RNA quadruplexes in human cells via an uncommon spectroscopic method
Abstract
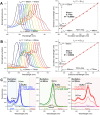
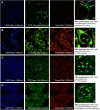
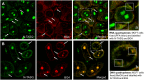
Guanine-rich DNA or RNA sequences can fold into higher-order, four-stranded structures termed quadruplexes that are suspected to play pivotal roles in cellular mechanisms including the control of the genome integrity and gene expression. However, the biological relevance of quadruplexes is still a matter of debate owing to the paucity of unbiased evidences of their existence in cells. Recent reports on quadruplex-specific antibodies and small-molecule fluorescent probes help dispel reservations and accumulating evidences now pointing towards the cellular relevance of quadruplexes. To better assess and comprehend their biology, developing new versatile tools to detect both DNA and RNA quadruplexes in cells is essential. We report here a smart fluorescent probe that allows for the simple detection of quadruplexes thanks to an uncommon spectroscopic mechanism known as the red-edge effect (REE). We demonstrate that this effect could open avenues to greatly enhance the ability to visualize both DNA and RNA quadruplexes in human cells, using simple protocols and fluorescence detection facilities.
Figures


Similar articles
-
Cellular Detection of G-Quadruplexes by Optical Imaging Methods.Curr Protoc Cell Biol. 2017 Sep 1;76:4.33.1-4.33.19. doi: 10.1002/cpcb.29. Curr Protoc Cell Biol. 2017. PMID: 28862343
-
Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe.Biochim Biophys Acta Gen Subj. 2018 May;1862(5):1101-1106. doi: 10.1016/j.bbagen.2018.01.022. Epub 2018 Feb 2. Biochim Biophys Acta Gen Subj. 2018. PMID: 29410183
-
Recent advances in fluorescent probes for G-quadruplex nucleic acids.Biochem Biophys Res Commun. 2020 Oct 8;531(1):18-24. doi: 10.1016/j.bbrc.2020.02.114. Epub 2020 Feb 25. Biochem Biophys Res Commun. 2020. PMID: 32111356 Review.
-
Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe.Nucleic Acids Res. 2018 Sep 6;46(15):7522-7532. doi: 10.1093/nar/gky665. Nucleic Acids Res. 2018. PMID: 30085206 Free PMC article.
-
NMR spectroscopy of G-quadruplexes.Methods. 2012 May;57(1):11-24. doi: 10.1016/j.ymeth.2012.05.003. Epub 2012 May 24. Methods. 2012. PMID: 22633887 Review.
Cited by
-
RNA G-quadruplexes: emerging mechanisms in disease.Nucleic Acids Res. 2017 Feb 28;45(4):1584-1595. doi: 10.1093/nar/gkw1280. Nucleic Acids Res. 2017. PMID: 28013268 Free PMC article.
-
Synthesis and investigation of quadruplex-DNA-binding, 9-O-substituted berberine derivatives.Beilstein J Org Chem. 2020 Nov 18;16:2795-2806. doi: 10.3762/bjoc.16.230. eCollection 2020. Beilstein J Org Chem. 2020. PMID: 33281983 Free PMC article.
-
Fluorescence Detection of DNA/RNA G-Quadruplexes (G4s) by Twice-as-Smart Ligands.ChemMedChem. 2025 Apr 1;20(7):e202400829. doi: 10.1002/cmdc.202400829. Epub 2025 Jan 15. ChemMedChem. 2025. PMID: 39714851 Free PMC article.
-
A tunable assay for modulators of genome-destabilizing DNA structures.Nucleic Acids Res. 2019 Jul 26;47(13):e73. doi: 10.1093/nar/gkz237. Nucleic Acids Res. 2019. PMID: 30949695 Free PMC article.
-
Small Molecule Fluorescent Probes for G- Quadruplex Visualization as Potential Cancer Theranostic Agents.Molecules. 2019 Feb 19;24(4):752. doi: 10.3390/molecules24040752. Molecules. 2019. PMID: 30791494 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

